In a previous post, I had critiqued Narasimhan et al 2019 for the inference that steppe ancestry is significantly related to modern Indian Brahmin groups, and hence provides evidence that this steppe ancestry is causal to the Indo Aryan languages.
Nevertheless, the fact that traditional custodians of liturgy in Sanskrit (Brahmins) tend to have more Steppe ancestry than is predicted by a simple ASI-ANI mixture model provides an an independent line of evidence—beyond the distinctive ancestry profile shared between South Asia and Bronze Eastern Europe mirroring the shared features of Indo-Iranian and Balto- Slavic languages (58)—for a Bronze Age Steppe the origin for South Asia’s Indo-European languages.
I gave 3 main reasons why this reasoning and analysis was faulty.
1. Correlation is not causation, Brahmin steppe can be enriched by way of intermarriage with steppe rich brides. It may not have anything to do with the steppe causing the formation of brahmin jAtis.
2. There are other brahmin jAtis in the same dataset from states other than UP and Bihar which do not show high Z scores as per the table given in the supplement. What explains that?
3. The main reason was that Steppe/Indus periphery ratio is a useless indicator in a 3 way model with Onge, steppe & Indus periphery. It would mean something only in a 2 way model between Steppe & Indus Periphery. I explain this with an example in my previous post, please read that if my reasoning is unclear.
Since then, I have studied this Z-Score table in the paper, and have found serious inconsistencies.
What is Z-Score?
The Z-score for each item in an array of numbers is simply the number of standard deviations that number is away from the mean of the array of numbers. Z>3 or Z<-3 (for eg. some may use 2, some 4) is indicative of that particular item being more than 3 standard deviations away from mean, at the extreme tail of the distribution, right and left end respectively. Therefore it is to be considered an outlier.
Z-Scores from the Narasimhan Paper
There are a couple of places where the paper defines the Z - Scores.
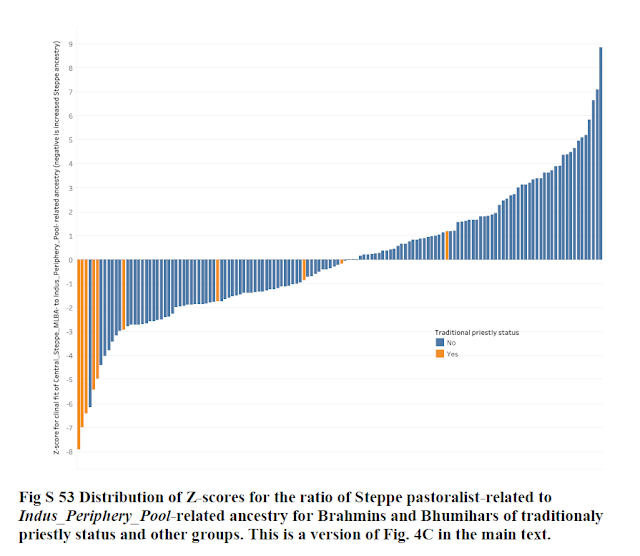
Our analysis of Steppe ancestry also identified six groupswith a highly elevated ratio of Central_ Steppe_MLBA– to Indus_Periphery_West–related ancestry compared with the expectation for the model at the Z < −4.5 level (Fig. 4). The strongest two signals were in Brahmin_Tiwari (Z = −7.9) and Bhumihar_Bihar (Z = −7.0). More generally, there is a notable enrichment in groups that consider themselves to be of traditionally priestlystatus: five of the six groups with Z < −4.5 were Brahmins or Bhumihars.
Read this - Distribution of Z Scores for the ratio of steppe pastoralist ancestry to indus pool related ancestry shows upto 8 standard deviation from the mean for brahmin groups as per the paper. THIS IS AN EXTRAORDINARY CLAIM.
I have been trying to reproduce these numbers for past 2 days, but just cannot arrive at such large numbers.
Z Scores have the property of having 0 mean and standard deviation as 1. But the Narasimhan paper has a standard deviation of 140 Z-Scores as 2.79 (indus periphery pool) and 2.06 (indus periphery west). There is clearly something seriously wrong in this part of the paper and its conclusion.
Methodology:
1. I copied the qpAdm coefficients, aposteriori hierarchical model coefficients and Z scores from Narasimhan et al 2019 excel supplement for all 140 populations into 2 excel sheets, one for Indus Periphery West and 1 for Indus periphery Pool models.
Indus Periphery West is sample I8726 who has the least AASI of all the Indus periphery samples from ShahrSokhta and Gonur. Indus Periphery Pool includes all 11 samples from both these locations, it has a higher AASI than I8726.
2. I made a column for Indus/steppe ratio in each sheet for all populations, and calculated the Z Scores for each population. Steppe/Inpe ratio will give same Z scores with sign reversed. I also used correlation on my calculated Z Score and the paper's Z-score using function '=correl(array1, array2)'.
3. Formula used for Z-Score: standardize(), which uses average() and stdev.p() as inputs for mean and standard deviation of the array of 140 ratios. ie '=standardize(x, mean, stdev.p)' for each of 140 x's.
The correlation of my Z Scores to the paper's Z Score for InPeWest is just 0.49, and 0.60 for InPePool.
Clearly this ratio is not what the paper used.
4. Next i calculated 140 Z Scores for the value 'Indus periphery MINUS steppe', using same formulae in step 3.
The correlation between this Z-Score and the paper's Z-Score was 0.9975 for InPeWest and 0.992 for InPe Pool. This seems to be what the paper used to calculate Z Scores - InPe MINUS Steppe ancestry %.
5. In none of these 2 Z Score arrays i calculated is the Z-Score for Brahmins >3 or <-3. They are all square in the meat of the normal curve.
As you can see, none of the Z-scores for Brahmins cross -3. So how did the paper get values of -7 and -8? I have tried various other combinations of the data, but none gave me high correlation of 99.8% with the paper's Z scores.
The paper's Z-scores look to be simply multipled 2-2.8 times of the actual Z-scores. I cannot fathom what calculation Narasimhan et al would have used to reach Z-scores of 7-8. It seems virtually impossible to me.
CONCLUSIONS:
1. The Z-scores based on which Narasimhan et al computed looks seriously flawed, and cannot be reproduced. The Z-scores do not have a standard deviation of 1, rather it's between 2-2.8. This means what the paper uses as ZScore isn't even Z-Score.
2. The Z-scores look artificially high, and cannot be reproduced.
3. The Z-scores seem to be calculated with Indus Periphery MINUS steppe ancestry as array input rather than ratios, which is a seemingly useless indicator.
4. The Z scores calculated based on either the ratio or the difference in indus/steppe ancestries do not cross the >3 threshold and are therefore not significant. It undermines one of the core arguments in the paper.
If any of you see some logic by which the paper's Z scores are correct, do let me know.
My workbook is below.Its downloadable.
References
Narasimhan VM, Patterson N, Moorjani P, et al. The formation of human populations in South and Central Asia. Science. 2019;365(6457):eaat7487. doi:10.1126/science.aat7487



20 comments:
If you can figure out how the paper got z-scores of 7 and 8, let me know. I'll give you a prize lol.
I was really surprised by 27% steppe in Kalash when Narashiman study came out because previous study claimed they were 50% steppe. What are your estimates?
Ror samples were not published yet when this Narasimhan study came out. How do you model Ror? they seem to have higher steppe admixture than Kalash, whats their admixture dates?
@Manu.V
I think this was the issue because back then the modelling did not take into account WSHGs. Previous modelling had Steppe eat up WSHG. But current modelling differentiates between the two.
@3rdacc
I don't think so, don't think any of them have it enough to cover 50%-45% steppe. Brahmin_Tiwari was 45% steppe in that study, which was second highest.
SIS2 has elevated levels of ANE in G25 but this is also not enough to cover such %.
People were making qpGraphs of Brahmins with 45%-47% steppe admixture with good z-scores before Narashian study.
It would be interesting to see how Rors and Haryanvi Jatts come out under Narasimhan model.
Jats are there in Narasimhan files. Check the workbook on this page
@vAsiSTha
Haryana Jatt is not in that, it's from different study. Haryana Jatt samples are more steppe-shifted for some reason, similar to Ror.
Both Ror and Haryanvi Jatt samples are from Pathak et al study. It was published after Narasimhan study. Sample is also labeled Jatt_Pathak in G25.
"SIS2 has elevated levels of ANE in G25 but this is also not enough to cover such %."
SIS2 having ANE is not a mistake.. Its definitely present, even in Irula.
Shahr I Sokhta BA2 Average
Fit 1.91
Ganj Dareh N 57.5
Simulated AASI by DMXX 32
RUS AfontovaGora3 8
Barcin N 2.5
GEO CHG 0
Kumsay EBA 0
Levant PPNB 0
Yes but i meant it has additional ANE outside of IranN component in SIS samples.
Yes, that's what I mean. The NW population existing in India around neolithic is around 88% IndiaN + 12% ANE/Tarim related.
Lets call that IndiaN1. Then Irula is 45%IndiaN1 + 55% AASI.
Z Scores have the property of having 0 mean and standard deviation as 1. But the Narasimhan paper has a standard deviation of 140 Z-Scores as 2.79 (indus periphery pool) and 2.06 (indus periphery west). There is clearly something seriously wrong in this part of the paper and its conclusion.
Mail sent to Narasimhan, Dr Reich and Dr Patterson
Dear Sirs,
Good day!
I have noticed a glaring error in the subsection of the South Central Asian paper regarding Z-Scores for the excess steppe ancestry of 140 modern Indian populations.
The paper claims
“Our analysis of Steppe ancestry also identified six groups with a highly elevated ratio of Central_ Steppe_MLBA– to Indus_Periphery_West–related ancestry compared with the expectation for the model at the Z < −4.5 level (Fig. 4). The strongest two signals were in Brahmin_Tiwari (Z = −7.9) and Bhumihar_Bihar (Z = −7.0). More generally, there is a notable enrichment in groups that consider themselves to be of traditionally priestly status: five of the six groups with Z < −4.5 were Brahmins or Bhumihars...”
I would like to inform you that the Z Score array (of 140 populations of model AHG + central_steppe+ Indus periphery pool) based on which this above conclusion has been drawn does not have standard deviation of 1, rather its standard deviation is 2.79. We all know that the std deviation of Z Scores should be 1.
This means that each Z Score has been artificially multiplied by a factor of 2.79. And if we reduce all 140 Z Scores by this factor, the Brahmin and Bhumihar Z Scores do not even cross threshold of 3 std deviations. The conclusions drawn based on this Z Scores are then also faulty and need to be retracted.
I trust the changes in the paper will be made accordingly.
You can read more about this on my blogpost https://a-genetics.blogspot.com/2022/01/scratching-head-narasimhan.html
Regards,
Vasistha, Manu V
"Jats are there in Narasimhan files. Check the workbook on this page"
No, haryanvi Jats are not there in narasimhan's paper. There are samples which are labelled as Sikh_jats which were taken from an earlier study(The earlier study collected those samples from a hospital via blood draws).
Here's the catch - From analysis of dozens of samples of sikh jats tested on private sites, hardly any of the sikh jat sample model like the ones in Narasimhan's paper. It was later found that those Narasimhan's Sikh jatts samples shared high IBD with Scheduled Castes so it seems that narasimhan's sikh jats were not really of 'pure' sikh jats .
On G25, sikh jats get higher steppe_MLBA than gangetic brahmins but less than Haryanvi jats. AASI
in both haryanvi and sikh jatts is roughly the same, it's just sikh jats get slightly higher BMAC and IVC periphery like ancestry than Haryanvi jats. So, it looks like - Punjabi
Sikh jatts are a mixture of Haryanvi jats + some Khatri-like group which makes absolute sense given the geography they inhabit.
"SIS2 has elevated levels of ANE in G25 but this is also not enough to cover such %."
@Manu, you are assuming that swapping Iran_N with SIS2 would a decrease of similar amount of steppe ancestry as much as the extra WSHG present in SIS2 , however, that assumes a linear model(simple addition/substraction) when trying to calculate the ancestries. This is not how their functions(mathematical transformations) work.
"People were making qpGraphs of Brahmins with 45%-47% steppe admixture with good z-scores before Narashian study."
Their is other thing you might be ignoring here and that is the ghost IndiaN population that Vasistha created. As per Vasistha's model, it is this IndianN population which provided a bulk of iranian related ancestry to Indus peripheries and also provided the southern ancestry to steppe eneolithic group. Since Indus Peripheries were used in narasimhan's model, it's highly possible that apart from excess WSHG, this latent IndianN ancestry in IVC-peripheries is what decreased the steppe_MLBA estimates (Earlier with Iran_N as source pop, Steppe_MLBA ancestry % took account of not only the genuine Steppe_MLBA ancestry but also the previously IndiaN and WSHG ancestry present in Indians)
makes sense. but genetic forums also have anecdotal evidence. looking for more good studies with more data on this - should be qpAdm, not G25 or ADMIXTURE.
"this latent IndianN ancestry in IVC-peripheries is what decreased the steppe_MLBA estimates (Earlier with Iran_N as source pop, Steppe_MLBA ancestry % took account of not only the genuine Steppe_MLBA ancestry but also the previously IndiaN and WSHG ancestry present in Indians)"
Good point.
I suggestion: "calibrate" your method with the replacement of the European NW population by Steppe people in the 3rd millennium BCE https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5973796/
@Anonymous
Thanks. Odd thing was both of those studies were from same Harvard team. Drastic difference in % was confusing as newbie.
@vAsiSTha
Are you suggesting steppe admixture in south asians is even lower than what is proposed by Narashiman et al?
Manu,
4-5% lesser for Swat valley iron age. For modern indians, I would go with narsimhan 2019 only. His reference populations used are decent. With tarim emba being published, things might change slightly.
I saw you wanted ss of R Khan's Twitter feed as he has blocked you. You can browse anyone's Twitter feed anonymously. Search for their Twitter account using a browser that you have not used for logging into Twitter.
Post a Comment