I received a reply from Harvard's Dr. David Reich to the mail I had sent (check previous post comments).
Hi Ashish,
The Z-scores are formal tests for model fit – appropriately calibrated by a block jackknife – and the tail of large Z-scores are simply reflecting the reality of model failure.
What the analyses are showing is that the “Indian Cline” is in fact best modeled as a two-dimensional cloud not a one-dimensional gradient: a mixture of three source populations that cannot be simplified to a mixture of two source populations as we had done in previous modeling for example in Reich et al. Nature 2019.
As such, what our enrichment analyses are showing is simply that the groups of traditionally priestly status tend to be enriched for the Steppe-related source population after controlling for the proportions from the other two.
Yours, David
I will address these points in this analysis post. This will be final evidence to disprove the confident Steppe--->Brahmin causality claim made in the study.
Let us make a new model to study excess steppe ancestry based on the qpAdm ancestries of the 140 modern populations studied in Narasimhan et al 2019 [1](the study). For the purpose of this analysis, we will ignore the bad qpAdm p-values in the study and assume that the models and ancestry estimates provided are accurate for all 140 populations, although some are clearly not.
We will assume that a 2 source ANI ASI model is accurate for modeling all modern South Asians.
ANI = a IVC + (1-a) Steppe
ASI = b AHG+ (1-b) IVC
where
ANI is Ancestral North Indian
ASI is Ancestral South Asian
a is ancestry coefficient of IVC ancestry (or Indus Periphery) in ANI
b is ancestry coefficient of AHG in ASI
0<=a,b<=1
Our main aim is to use the qpAdm coefficients for each population, and by keeping the AHG and IVC ancestries the same as the qpAdm model (thereby fixing the amount of steppe, which will be different than the qpAdm model) but varying a,b in such a fashion that the overall errors in the 140 groups are minimized.
let G be one of the 140 modern groups.
G = e.ANI + f.ASI; 0<=e,f<=1 & e+f=1
G = ea.IVCN + eb.Steppe + fc.IVCS + fd.AHG
ea + fc = qpAdm IVC estimate for group G
fd = qpAdm AHG estimate for group G
IVCN = IVC ancestry type from ANI part
IVCS =IVC ancestry type from ASI part
For easier understanding, let me explain it with kalash.
qpAdm for Kalash: 0.123 AHG + .599IVC + 0.278 Steppe
we can write it as
If both these ratios are 0.4, it would mean
ASI = 71.24% AHG + 28.76%IVC
and ANI = 71.24%IVC + 28.76% Steppe
and Kalash would become
0.123 AHG + 0.0492 IVCS + 0.5498 IVCN + 0.2199 Steppe.
We can see that the steppe component of 0.2199 is less than the qpAdm estimate of 0.278. ie the model predicts that the ANI for Kalash had a higher steppe component than what we chose. This difference of 0.0581 is the fit error for this group for this particular choice of ratios.
Fitting all 140 Groups with the ANI-ASI model
For a particular set of ratios (1-a)/a and (1-b)/b, we will get a set of 140 fit errors for 140 different groups. We can now vary both the ratios and come to a final choice of ratios at which the sum squared of the 140 fit errors is minimized. We also place the constraint that none of the 4 ancestry coefficients can be negative.
This can be done via Excel VBA macro, or more simply by using the solver plugin in excel.
The error minimizing algorithm gives the result for IVC/AHG ratio in ASI as 0.421 and that of Steppe/IVC in ANI as 0.372. ie ANI = 72.9% IVC + 27.1% Steppe and ASI = 70.36% AHG and 29.64%.
The excess steppe error ranges from -0.13 to 0.18 steppe % points with a mean of 0.0015 and a median of -0.0037 and a standard deviation of 0.0527. mean and std dev estimated from 5000 Monte Carlo random simulation runs of picking errors of 15 random groups each time.
Converting these error scores to Z-Scores by standardizing, results in a Z-Score range of -2.41 to 2.18, with mean 0 and std dev 1 - confirming that it is indeed Standard Normal Z distribution.
 |
| ANI ASI Model shows significant outliers from the cline |
A Working 3 Source model for 140 modern Indian groups
eg.
qpAdm estimates for Kalash: 0.123 AHG + 0.599IVC + 0.278 Steppe
where g is the proportion of AHG for Kalash which is part of ASI, 0<g<1. there are 140 of these - g1 to g140.
r1 = ratio of IVC to AHG in ASI; common for all 140 groups
r2 = ratio of IVC to AHG in AI; common for all 140 groups
r3 = ratio of Steppe to IVC in ANI; common to all 140 groups
The coefficient we get for Steppe is compared to the qpAdm estimate and the difference is the error. Our error minimization algorithm will vary these 143 parameters such that the sum squared of the 140 errors is minimized.
We use the Excel Solver plugin for this. One additional constraint is that none of the 6 coefficients should be negative.
RESULT
After the error minimization, we get a working model.
The mean of the error is 0.0001, the median is 0.0000, the standard deviation is 0.0014. The range of the error is from -0.0014 to max 0.017. The Z-Scores for model fit (error/SE) range from -0.0969 to 1.30, giving proof of successful fits for all 140 populations.
The error range of -0.13 to 0.18 in the best fitting ANI ASI model is reduced to -0.0014 to 0.017, and the standard deviation is reduced from >0.05 to 0.0014.
This is expected because we are already assuming that a 3 source model given by qpAdm (AHG + IVC + Steppe) is accepted, therefore any 3-way combination of these 3 underlying ancestries will also give a successful fit. However, the 3 sources I have chosen have a chance of providing more historical information as a proximal model.
From the output of the fitted model we get:
ANI = 64% IVC + 36% Steppe
ASI = 99% AHG + 1% IVC
AI = 61% IVC + 39% AHG
and all 140 groups as a combination of these 3 ancestries.
The PCA of all 140 groups with these 3 proximal sources is below
THE BRAHMIN OUTLIER GROUPS
As per the 3 source model above, none of the 4 outliers - Bhumihar_Bihar, Brahmin_UP, Brahmin_Tiwari, and Brahmin_Nepal, are in need of excess steppe ancestry over and above what is provided by ANI. In other words; Nepal, UP, Chattisgarh & Bihar Brahmins DO NOT NEED A SEPARATE ANI SOURCE WITH HIGHER STEPPE/IVC ANCESTRY RATIO.
The excess Steppe needed in the failed model was actually a false signal, what is needed in these 4 outlier groups is an ancestry source that has excess ASI(AHG) and almost nil IVC on top of ANI as well. This is exactly the point that I had made in my Part 1 critique, that eastern Indians have excess AHG ancestry as well, which is driving this false signal of excess steppe requirement in the failed ANI ASI model.
If it were not for this high AHG source requirement specific to these 4 groups, they would not have needed a high Steppe/IVC ANI source to model them.
An important corollary is this: If a set of populations can only be modeled as a 3-source admixture, then only 1 ancestry (or its ratio with any 2nd) cannot be the only explanation for a 2-source model failure. By mathematical certainty, at least 2 sources / 2 ratios will explain the deviation from the 2 source model.
How can it be? How can this ANI group meet with almost pure original AHG (also called AASI) ancestry? It appears that eastern India still has populations with the highest AHG like ancestry even in modern times, even more so than the southern Dravidian Paniya & Irula tribals. The Birhor tribals of Jharkhand, Bihar, Chattisgarh are Austroasiatic-speaking hunter-gatherers. They possess even greater AASI/AHG ancestry than Paniya, mixed with SE Asian ancestry, and there is some evidence to suggest that even the little IVC like ancestry in Birhor is of recent admixture (DATES: S1 Shahr_Sokhta_BA2, S2 ONG.SG, Target BIR.SG: Output 23.77 +- 8.65 generations, 1092CE - 1576CE)
This is additional evidence to the almost consensus hypothesis that AASI entered India from the East/Southeast of India in paleolithic times.
This can explain how these 4 Brahmin groups from east India picked up AASI/Austroasiatic ancestry. At some point, they must have intermarried locals from the region that they settled in. These Brahmin groups are just showing the adaptation to their region, just like other groups show their adaptation to their specific regions. Another line of evidence to prove this hypothesis is that eastern Indo Aryan languages show Munda language substrate which is absent from western IA languages, hinting at IA speaking groups changing the language of the eastern Indian region where Munda people lived.[2][3][4] |
| UP Brahmins from Mondal data can be informally modeled as Kalash + Birhor with OK fit. |
qpAdm for Birhor: P-value 0.0003 (best fit i got): 0.69 Onge + 0.175 IndusPWest + 0.138 Dai
PCA Based on Language
Then is there a way to check if this ANI ancestry has any special relation to Language and religion?
 |
| A significant difference in the ANI component between Brahmins and non Brahmins |
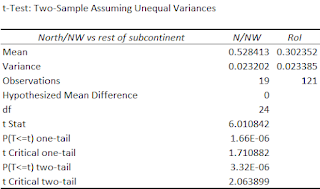 |
| A significant difference in ANI between NW India/Pak and the Rest of India |
N/NW India = Groups from Pakistan, Delhi, Haryana, J&K, Punjab, Rajasthan.
So what Null hypothesis should we test?
 |
| The null hypothesis of equal ANI in Brahmins & NW India cannot be rejected |
Bear in mind that the Brahmin groups don't include lower ANI groups like Marathi, Tamil, Gujarati brahmins, etc, whereas the NW groups don't include higher ANI groups like Ror, Punjab_Jatt, Kamboj, etc. For sure, we could be missing the other side of the same coin too, but I don't think there are any other Brahmin groups with more ANI than UP brahmins. The more data we have the better the statistical power of the tests will be.
CONCLUSION
Nevertheless, the fact that traditional custodians of liturgy in Sanskrit (Brahmins) tend to have more Steppe ancestry than is predicted by a simple ASI-ANI mixture model provides an independent line of evidence—beyond the distinctive ancestry profile shared between South Asia and Bronze Eastern Europe mirroring the shared features of Indo-Iranian and Balto- Slavic languages (58)—for a Bronze Age Steppe origin for South Asia’s Indo-European languages.
References
Downloadable workbook

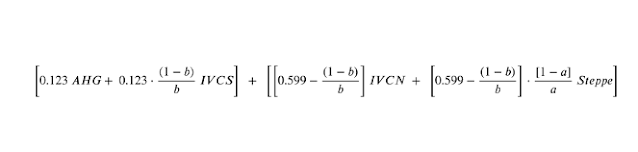



38 comments:
If Kalash are 21% then UP Brahmin/Brahmin_Tiwari would be close to 15-16%? Ror would be bit over 27%?
Lazaridis study Brahmin_Tiwari 45% steppe, with IranN, EMBA steppe, Onge model.
Narasimhan study Brahmin_Tiwari 25% steppe, with new SIS2 samples, MLBA steppe, AASI model.
Under your model Brahmin_Tiwari will be close to 15% steppe and Kalash are 21% steppe? with your proposed "IndiaN1" ghost?
I don't say kalash = 21% steppe. In this analysis I'm not challenging the papers qpAdm coefficients. That might come later. Although by and large those are good models, just low pvalues for many populations.
Tiwari 45% steppe is impossibly high. Those are older models, forget them.
No Indian population will have more than 35% steppe_mlba.
The three way model makes more sense because actual admixture sources can never be that simplistic. I also think that zombie populations like Srilanka's vedda may harbour Iran like component than Onge which is more eastern.
The paper itself rejects a 2 source model and goes on to show a 3 source model, however never does the excess steppe analysis on the 3 source model. Dr Reich in the mail also admits that 2 source model is a failure.
Yes, i think east india will have more AASI like populations that South.
Also it can be expected that any clines in the subcontinent would be dominated more on an east west axis vs a north south one simply because the gangetic plains and Himalayan foothills would have always fielded higher populations with connections further east, vs that of peninsular India which is bounded by ocean.
@Vasistha
Does the ENA affinity in ANE can be mediated from AASI like source props?
@ganesh
doubtful. its some sort of pre-Onge & AASI east eurasian.
@vasistha
The reply from David Reich is suggestive. Those high z-scores are only the artifact of a failed model - that they use this to derive a conclusion without rational basis is evidence of "intent to mislead" (at the very least).
Coming to your proposed three group model, it is interesting to see that a lot of "AI" type populations still exist in Modern South India today -
Coorghis (Kodavas), Agnikula Kshatriyas (Andhra), Vysysas (Andhra), Reddy (Telangana). They all speak Dravidian languages and are Sanskritised in customs (Kodavas are exception).
@bruin
I don't think it's intent to mislead. I think it's confirmation bias. They didn't drill deeper when the results showed the outcome in their bias.
@vAsiSTha
35% for Rors will be close to Narashimhan model then.
In previous posts comment you said steppe will be "4-5% lesser for Swat valley iron age" and you also mentioned "With tarim emba being published, things might change slightly."
I think that swat sample had 19% steppe in Narashimhan, right?
Including Tarim_emba as source for Swat G25 (informal) reduces sintashta ancestry by 4-5% to around 10-15% average.
also worth noting that Sintashta is western_steppe_mlba, but narasimhan uses central_steppe_mlba in swat model which is sintashta + minor excess wshg. this allows the steppe component to again expand by 3-4% in Swat, as some of the tarim_emba is subsumed by the central_steppe.
@vasistha
Understood what you meant. Coming to your three source model -
I suppose that the limiting factor is the availability of of only one Bronze Age aDNA cline (IVC) in India. It is absolutely necessary to have 3 restricting equations in order to have a solution for three unknown variables.
We need a BA Magadhan Cline from Bihar and a BA Deccan cline from south of the Vindhyas.
The BA Magadhan Cline is going to be pretty interesting as that is the home of the Sramana religions as opposed to Vedic religion. And there is extensive academic consensus that Sramana philosophies predate the Vedas.
The BA Deccan cline is going to tell us if there was ever such a thing as pure AASI in the Bronze Age. I doubt this!
> And there is extensive academic consensus that Sramana philosophies predate the Vedas.
This is because they say Vedas are max 1500 BCE, and can't be any older. There is no evidence showing whether one is older than the other.
? I don't believe the sramana consensus. It certainly isn't based on the Munda like people of eastern India before the region got aryanised.
@Vasistha @3rdacc
The Vedas (Book 3) explicitly mention that regions downstream on the Ganges are "Anarya" - they don't fall into the continuum of rituals of fire worship and Agni-sacrifice. Even though it is clear that they use "Anarya" in a religious sense, they seem to them as Others.
In time they were Aryanised - but they retained the primacy of the King over the Brahman. We see two major differences between the Gangetic tradition (Ramayan) and the Indraprastha tradition (Vedas, Mahabharata). The King/Prince himself is the Perfect Godhead (Avatara) in Ramayan but in the Mahabharata - the focus is on the follies of the kings and princes. The Godhead (Krishna) steps in to resolve the moral dilemmas.
Historically we have a strange parallel - Chanakya travels from Takshashila (NorthWest) to Pataliputra to help the Mauryas overthrow the Nandas. He could never become a King, but only the power behind the King.
Excess Steppes in Gangetic Brahmin groups is not so strange after all!
I am genuinely curious to know what was the autosomal make-up of Bronze Age Bihar at the time of IVC. It will illuminate the current imbroglio greatly.
Bronze age Bihar will have Munda like tribals
Interesting, so since I am more interested in ultimate ancestry components so to speak you'd say that South Asians can be modeled as something related to IVC, something related to Steppe and something related to Onge?
In respect to this Narasimhan et al (2019) claimed that the Iranian-related ancestry found in the Harappan samples is not quite Iran_N but a diverged version of it. Iosif Lazaridis claimed that it is Iran_N + ANE/ENA. Which model would you go with?
Finally, the impression I get from this model is that IVC-type pops stayed in northern India quite a long time and only later really mixed with AHG-type groups. In that sense, there was more separation between the people living in north and south India at the time possibly with low gene flow between them?
> The Vedas (Book 3) explicitly mention that regions downstream on the Ganges are "Anarya" - they don't fall into the continuum of rituals of fire worship and Agni-sacrifice. Even though it is clear that they use "Anarya" in a religious sense, they seem to them as Others.
This is certainly true. The Arya "center" was the whole Punjab-Haryana region.
> In time they were Aryanised - but they retained the primacy of the King over the Brahman. We see two major differences between the Gangetic tradition (Ramayan) and the Indraprastha tradition (Vedas, Mahabharata). The King/Prince himself is the Perfect Godhead (Avatara) in Ramayan but in the Mahabharata - the focus is on the follies of the kings and princes. The Godhead (Krishna) steps in to resolve the moral dilemmas.
never really heard of this before.
"Interesting, so since I am more interested in ultimate ancestry components so to speak you'd say that South Asians can be modeled as something related to IVC, something related to Steppe and something related to Onge?"
combination of 3 ancestries
ANI - 65%IVCWest + 35% Steppe
ASI - 100% AHG
AI - 60% IVC west + 40% AHG
2 caveats here
1. Based on only the 140 modern groups listed in Narasimhan supplement excel, not others.
2. assumption is that the qpAdm models in study are accurate models, which theyre not as many p-values are bad.
"In respect to this Narasimhan et al (2019) claimed that the Iranian-related ancestry found in the Harappan samples is not quite Iran_N but a diverged version of it. Iosif Lazaridis claimed that it is Iran_N + ANE/ENA. Which model would you go with?"
I go with both. The non AHG component is IndiaN + ANE/Tarim like. around 10-15% ANE on avg.
"Finally, the impression I get from this model is that IVC-type pops stayed in northern India quite a long time and only later really mixed with AHG-type groups. In that sense, there was more separation between the people living in north and south India at the time possibly with low gene flow between them?"
I see quite high geneflow between south and north given that IA type ancestry is very widespread (except for east indian tribal groups)
Please, analyze Britannia's Steppe admixture. I don't see any meaningful cultural change from the 4th mil BCE to half 2nd mil BCE, which is when Celtic continuity seems to be clear. But it is said that there was replacement at the mid 3rd millennium by steppe people. But there is continuity in the continent with the Basques. Also, Etruscans seems to be kind of invisible.
Maybe the Steppe signature is overblown and if you analyze that, that should decrease even more the presence of that in India, since Britannia is the most extreme example.
I will Daniel, soon.
I2872 from RoopkundA is a woman who is most likely a UP/Bihar Brahmin. Her ancestry lays on the same ANI-AHG cline as the 4 Brahmin groups, only she lies slightly closer to ANI than moderns.
@Vasistha
"2. assumption is that the qpAdm models in study are accurate models, which theyre not as many p-values are bad."
I saw a post by Davidski on this topic at some point, apparently Onge are not a good proxy for the AHG ancestry in South Asians (too drifted/is AASI Onge+X or Onge AASI+X?not sure)
What else would cause such low p-values (which I see affect all groups, rather than just say potentially Austroasiatic-influneced ones)
"I go with both. The non AHG component is IndiaN + ANE/Tarim like. around 10-15% ANE on avg."
So in this case the ENA in India_N is a more northern ENA component rather than AHG?
"I see quite high geneflow between south and north given that IA type ancestry is very widespread (except for east indian tribal groups)"
Today sure, but it seems during IVC times this was not the case as much. In the Swat samples for example it seems AHG increases over time.
"Onge are not a good proxy for the AHG ancestry in South Asians (too drifted/is AASI Onge+X or Onge AASI+X?not sure)"
In qpAdm its a good proxy as its insensitive to drift. in g25 its not at all a good proxy. AASI forms a clade with AHG, no extra admixtures.
"So in this case the ENA in India_N is a more northern ENA component rather than AHG?"
Yes. IndiaN1 = 0.88IndiaN + 0.12ANE is my assumption. Although Narasimhan believes it could have come through AHG also.
from the supplement
We particularly focused on the case where the proportion of AHG-related ancestry was zero, in which case the proportions of Ganj_Dareh_N- and WSHG-related ancestry are 89 ± 10% and 11 ± 10% respectively. Similarly, on the other end of the cline, the proportions of AHG and WSHG-related ancestry (setting Ganj_Dareh_N-related ancestry to zero) are 97 ± 10% and 3 ± 10% respectively. While the confidence intervals for WSHG ancestry at both ends of the cline overlap 0%, this does not imply that the Indus Periphery Cline individual themselves had no WSHG-related ancestry (to the contrary, the qpAdm analysis proves that they did). What these numbers instead show is that we cannot determine what end of the cline contributed the WSHG-related ancestry.
"What else would cause such low p-values (which I see affect all groups, rather than just say potentially Austroasiatic-influneced ones)"
Presence of additional east asian and/or ANE ancestry. i have detected many such samples in my roopkund post. you can see their qpAdm results there, for all 23 samples.
*Although Narasimhan believes it could have come through AHG also.
Narsimhan says the ANE could have come along with AHG in indus periphery.
I doubt it. darra-i-kur 2500bce afghanistan has no AHG but enough ANE.
"AASI forms a clade with AHG, no extra admixtures."
That makes sense, and would you agree that in deep perspective AASI/AHG form a clade with Tianyuan or do they have other types of admixtures? Most papers model them as descending from the same pop but not sure.
This part from Shinde et al (2019) is quite interesting:
"The only fitting two-way models were mixtures of a group related to herders from the western Zagros mountains of Iran, and also to either Andamanese Hunter-Gatherers (73 ± 6% Iranian-related ancestry; p=0.103 for overall model fit) or East Siberian Hunter-Gatherers (63 ± 6% Iranian-related ancestry; p=0.24 (the fact that the latter two populations both fit reflects that they have the same phylogenetic relationship to the non-West Eurasian-related component of I6113 likely due to shared ancestry deeply in time)"
What would you make of it? Imo, it likely hints at both ancestries that is AHG and East Asian-related existing in IVC.
Iran_N was modeled as part Onge in the Dudzuana paper but it was a kind of lazy model since I doubt Iran_N is part Mbuti. And then CHG was part Tianyuan, so anyways I think a more northern source is more likely since ANE is involved than AHG in Afghanistan or Iran.
"Presence of additional east asian and/or ANE ancestry. i have detected many such samples in my roopkund post. you can see their qpAdm results there, for all 23 samples."
Interesting, skimming through them it seems like the admixture is proper East Asian. I wonder whether Chokhopani-type pops have anything to do with it. That's where the upcoming aDNA from Nagaland will likely be handy.
@gamerz_j
Shinde et. al. 2019 says following,
"
If one of these population fits, it does not mean it
is the true source; instead, it means that it and the true source
population are consistent with descending without mixture
from the same homogeneous ancestral population that poten tially lived thousands of years before"
I think it really points out the deep phylogeny relation between AASI, AHG and ESHG, it does not does not mean Harapans had AHG and East Asian ancestry directly.
@gamerjZ
"That makes sense, and would you agree that in deep perspective AASI/AHG form a clade with Tianyuan"
Yes, they do. no admixtures.
"What would you make of it? Imo, it likely hints at both ancestries that is AHG and East Asian-related existing in IVC."
It's just AASI. Rakhigarhi sample has low SNP count, hence multiple accepted models.
@Ganesh
Yeah that seems to be the case actually, like Vasistha pointed out Rakhigarhi sample is low coverage so probably that's mainly the reason both models could work. As well as the shared ENA ancestry in ESHG and AHG/AASI.
@Vasistha
If/when you have some time could you please run a qpAdm on Onge using Tianyuan, Ustishim and Mbuti or/and Kostenki (would that work?) as sources? Mostly to just have a handy reference about the phylogeny of Onge and because in some papers they seem to have some sort of relationship with Ustishim (and not just them but southern ENA in general)
Or if you have any stats handy to that effect they would also be welcome!
I have done many qpgraph posts with Mbuti, tianyuan onge kostenki MA1 etc.. onge just forms a clade with tianyuan, and another with AASI.
@Vasistha
Yes, I am just wondering if the Onge show the same thing in qpAdm. Even if you have tried it in the past or something.
Reason I am asking is because one post you had on Eurogenes had Onge slightly Mbuti (something like 2% was it) and I can't find it atm, I don't remember if that was using qpGraph or qpAdm.
That would be just noise i guess
"No Indian population will have more than 35% steppe_mlba."
@Vashistha,
Some outliers samples from hindu jats and rors will touch 37%-38% Steppe_MLBA despite using an 'India_N'+WSHG enriched BMAC source as one of the source pops as shown in G25 models.
"but I don't think there are any other Brahmin groups with more ANI than UP brahmins."
I think some Rajasthani, punjabi and gujarati brahmins will indeed have more ANI(IVC+Steppe) than UP brahmins based on samples i have seen.
Ani is not just IVC + steppe. Or even (1- AHG).
It's a component with a certain fixed steppe:ivc ratio. The excess ivc apart from this goes into AI component (as per my model) or ASI (if 2 way ANI ASI model). If steppe is higher, then ANi is higher and vice versa, as ANI is only steppe source.
In general, we can drop such models because it gets confusing. Just use ivc, steppe, AHG, Wshg/tarim, east asian. This model also doesn't consider later steppe intrusion as a possibility, so it's wrong anyway.
Yea, jats and rors weren't part of the 140 groups in the study so this model of mine (or Narasimhan) is not made to fit them.
Thanx vasistha for ANI part clarification.
"Just use ivc, steppe, AHG, Wshg/tarim, east asian."
Why not use a turanian shifted and ANF depressed BMAC sources like Parkhai MBA (or Dzharkhutan_BA) too ? Its really impossible to bypass the BMAC and i have been told that if one adds Parkhai_MBA and SIS_BA1(one of the more west shifted IVCp) as sources, many north-west and north indian pops seems to prefer Parkhai_MBA over SIS_BA1. Correct me if i am wrong but narasimhan rejected BMAC as the source because of taking whole population of BMAC as one single cluster, right !
I'll check parkhai mba..
Iirc, narsimhan either used Gonur or bustan as proxy for BMAC
Hi, I've heard some claims that R1a/R1b related genes are responsible fair skin among Indians and that there is a strong correlation between them. Can you shed some light on this?
Hello Ashish
Recently I sent an email to vashisth if iam right u both are same person. I have found an interesting athrophometric which can be linked to genetics for identifying brahmins. I fact based on my study brahmins would cluster more to north west people , middle east and minimal to western, eastern europeans. I can provide evidences based on my study.
Post a Comment