Came upon two linguistic papers which clarify the relationship of Aryan languages of India with the other old language families, namely Dravidian & Munda families.
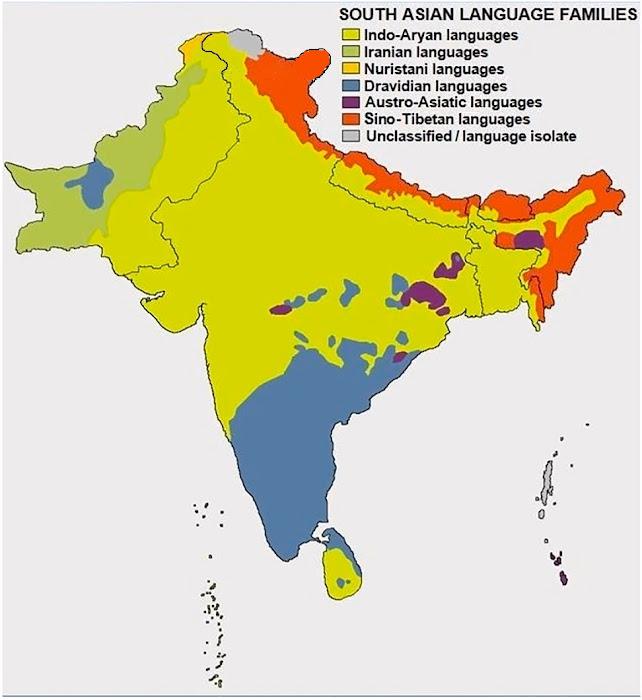 |
| Language Family map of India |
I recently wrote a post in which my analysis showed that The same ancestors which provided iranian like ancestry to Irula tribals also provided ancestry to Steppe Eneolithic as well as South Central Asia (Sarazm aDna etc). One can read the post here.
For feedback as well as spreading the post, I posted the link to a popular DNA & population genomics forum - Anthrogenica. Quite unexpectedly, immediately after, I was suspended from Anthrogenica with no reason or message whatsoever. I always knew it is a Kurganist bastion, but never quite expected this level of censorship to opposing ideas. I am glad though, starting this blog is now worth it. Also, credit to Davidski at Eurogenes blog for allowing me on his blog comments section even with my opposing views.
I came to know that the Moderator who banned me is a handle named Coldmountains, a handle who I have in the past ridiculed quite a lot (in Eurogenes comment section) for not finding a single R-L657 indian y haplogroup in the steppe since 2015. Poor guy comes empty handed after each successive paper when new samples from the steppe are published. His search still goes on. Meanwhile the only L657+ sample we have so far in aDna is from Roopkund lake India 800CE.
The link to my Anthrogenica thread is here. Please register and show Anthrogenica some love in this thread and elsewhere. The moderator clique there is in an echochamber and needs some awakening.
Anyway, let move on to the criticism of my post. There is just one, and sadly i couldn't reply because I was banned. Hence this post.
Kale on 25-Nov-2021 wrote
Kotias is a pseudo-haploid sample > That means rather than having two different sets of chromosomes like a real person, it is treated as having two exactly identical sets > That means the drift going to itself it going to be crazy high > If you have an edge coming out of an artificially crazy high drift, the percentage contribution has to be artificially crazy small to avoid overfitting.
This graph is completely uninformative until structured properly.
Kale is absolutely wrong here. The pseudo-haploid* samples do not cause artificial high drift edges, rather, the artificially high drift is due to just 1 sample in the label because of which heterozygosity cannot be computed for the label. This problem is solved by using 2 samples in the label even if samples are pseudo-haploid. This is not a problem for .DG samples as these are diploid genomes and allow for heterozygous calls.
This is exactly what I have done in the graph below (later). I lumped Satsurblia & Kotias into 1 label known as CHG. I will show that my conclusion does not change.
Proof of my claim is from the programmers of Admixtools in their qpGraph readme pasted below. Should have been basic reading right?
Genotypes are expected to be pseudo-haploid -- 2 samples at least per population or drift lengths on leaves are not meaningful.
As far as edges coming out of artificially high drifts are concerned, sister clades of Kotias also did not help Kales case. See, i spent weeks on the model trying every possibility. Them not being able to read the graph is not my problem.
Below I will paste my new qpGraph for Steppe Eneolithic which follows these principles and should be acceptable to the Kurganists as well.
Please click on the graph for high res mobile view. On Desktop download image and zoom in a zoomable picture viewer.
DISCUSSION
After correcting all criticisms, the need for IndiaN component in Steppe eneolithic does not go away. I again prove that the same ancestors who ultimately provided ancestry to Steppe Eneolithic in 5th mil BCE also provided ancestry to Irula tribe (and by extension most of indian groups). The minute criticisms which Kurganists come up with are immaterial now, because of course they will come up with them. So far, they have been busy denying even Iranian inflow into steppe (its a mater of purity of course!), so to accept South/ SC Asian origin is a different matter altogether.
 |
| Representative example of a user's data being merged into Eigenstrat format for Admixtools |

Domestication of horses fundamentally transformed long-range mobility and warfare. However, modern domesticated breeds do not descend from the earliest domestic horse lineage associated with archaeological evidence of bridling, milking and corralling at Botai, Central Asia around 3500 BC. Other longstanding candidate regions for horse domestication, such as Iberia and Anatolia, have also recently been challenged. Thus, the genetic, geographic and temporal origins of modern domestic horses have remained unknown. Here we pinpoint the Western Eurasian steppes, especially the lower Volga-Don region, as the homeland of modern domestic horses. Furthermore, we map the population changes accompanying domestication from 273 ancient horse genomes. This reveals that modern domestic horses ultimately replaced almost all other local populations as they expanded rapidly across Eurasia from about 2000 BC, synchronously with equestrian material culture, including Sintashta spoke-wheeled chariots. We find that equestrianism involved strong selection for critical locomotor and behavioural adaptations at the GSDMC and ZFPM1 genes. Our results reject the commonly held association between horseback riding and the massive expansion of Yamnaya steppe pastoralists into Europe around 3000 BC driving the spread of Indo-European languages. This contrasts with the scenario in Asia where Indo-Iranian languages, chariots and horses spread together, following the early second millennium BC Sintashta culture
 qpAdm is a statistical tool for studying the ancestry of populations with histories that involve admixture between two or more source populations. I hope this tutorial helps people interested in doing own research as qpAdm validated models which are constructed well provide a high standard of evidence to the model.
qpAdm is a statistical tool for studying the ancestry of populations with histories that involve admixture between two or more source populations. I hope this tutorial helps people interested in doing own research as qpAdm validated models which are constructed well provide a high standard of evidence to the model.North-West Indian population groups had a total of 55.86% of samples characterised as belonging to South Asian ancestry haplogroups (M, U2, U4), followed by West Eurasian (40.18%, H, HV, I, J, K, N, R, R0, T, U1a, U5a, U7, U8a, W, X) and East Asian (3.96%, A, B, C, D, F, G ) (Fig. 1).
The below analysis is mine, after studying the raw data in the paper.
I will make 3 separate posts on how to install Admixtools, run qpAdm and run qpGraph respectively. This first blog post will detail steps to successfully install Admixtools by Harvard lab.
Admixtools cannot be run from a Windows system. If you have a Windows or MacOS Host system, install Oracle VirtualBox from here. Its free.
Install any Guest linux system on this VBox. I use OpenSuse Leap v15.1 for no particular reason apart from that I'm used to it. The virtual image for installation can be downloaded for free from here.
Once this is done, install the guest Linux system on the VBOX. This video below can guide you.
I am working on some qpGraphs to figure out the different Iran farmer like ancestries in Ganj Dareh, CHG, TepeAbdulHosein, Hotu cave, Wezmeh cave, SC Asia (Sarazm) and India etc. Its taking longer than I expected because of many populations adding complexity.
So I thought i would use the graphs already made to shed some light on some Eurasian samples. When I publish qpGraphs please keep some things in mind while analyzing.
From Hallast, P., Agdzhoyan, A., Balanovsky, O. et al. A Southeast Asian origin for present-day non-African human Y chromosomes. Hum Genet 140, 299–307 (2021). https://doi.org/10.1007/s00439-020-02204-9
Here, we show that phylogenetic analyses of haplogroup C, D and FT sequences, including very rare deep-rooting lineages, together with phylogeographic analyses of ancient and present-day non-African Y chromosomes, all point to East/Southeast Asia as the origin 50,000–55,000 years ago of all known surviving non-African male lineages (apart from recent migrants). This observation contrasts with the expectation of a West Eurasian origin predicted by a simple model of expansion from a source near Africa, and can be interpreted as resulting from extensive genetic drift in the initial population or replacement of early western Y lineages from the east, thus informing and constraining models of the initial expansion.