I have been working on this project for 3 weeks, and results are nothing less than spectacular! But for those who follow my comments on other blogs, this is hardly surprisng.
In 2019, 3 very important human remain samples from the Caucasus region were published
1. 2 of these samples were from a site called Progress II, and 1 from a site known as Vonjucka. Both these sites are just north of the caucasus mountains in now Southern Russia. The carbon dating of these samples shows that the oldest lived around 4900 BCE and the newest around 4200 BCE.
What is important about these samples is that they possess the oldest autosomal aDna which corresponds to the genetic components which we now call "steppe aDna". This autosomal ancestry is now widespread from western europe to central asia and south asia to various degrees. These samples are now labeled as Steppe_Eneolithic or Steppe_En, and I shall also address them as such below.
 |
| Location of the Steppe Eneolithic Samples |
Steppe_En gives a large chunk of ancestry to the Yamnaya culture in steppe and Corded Ware culture of western Europe, and is a contender for the population which spread the IE language to Europe.
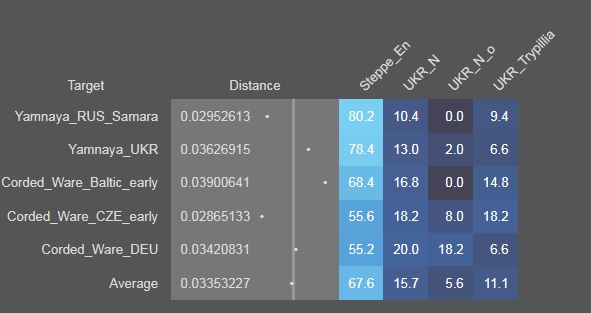 |
| Rough estimate using G25 and Vahaduo shows 55-80% Steppe_En ancestry in Corded ware and Yamnaya |
Now that we have established the importance of Steppe_En, lets understand what ancestry Steppe_En itself is made of. The 2019 Caucasus paper1 states this about Steppe Eneolithic.
North of the Caucasus, Eneolithic and BA individuals from the Samara region (5200–4000 BCE) carry an equal mixture of EHG- and CHG/Iranian ancestry, so-called ‘steppe ancestry’ that eventually spread further west, where it contributed substantially to present-day Europeans, and east to the Altai region as well as to South Asia.
Where EHG = EastEuropean Hunter Gatherers, represented by samples from Karelia & Samara in Russia around 6000 BCE. CHG = Caucasus Hunter Gatherers, represented by samples from Satsurblia & Kotias cave in Georgia dated to around 11k BCE and 7500 BCE respectively.
As you can see from quote, CHG/Iran is used, where Iran is meant to be Ganj_Dareh_neolithic samples ancestry from Zagros mountains in West Iran. I shall use the label IranN in this post for these samples. The lack of anceint samples from west asia and caucasus makes it difficult to identify whether the Steppe_En ancestry is from CHG or Iran
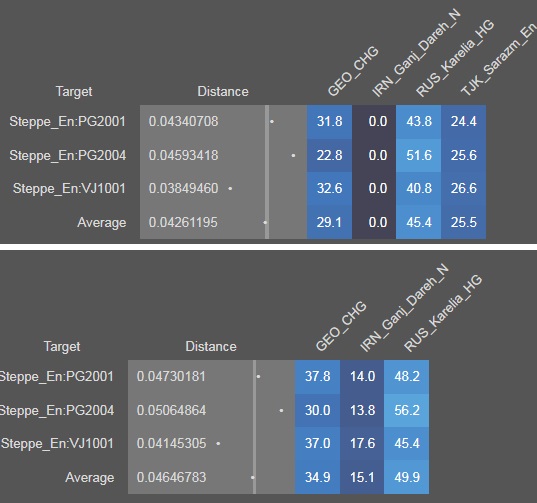
Lets see what G25 and Vahaduo says
As we can see, a south central population (Sarazm) is chosen over IranN as input and with Sarazm the distance reduces significantly. Although the overall distance at 4.26% is still bad and shows that we are missing something. But we have already started seeing some connection with SC Asia.
I ran qpAdm with the EHG + CHG model, but the model fails because the actual samples show more affinity to IranN but the model doesn't have it. Output is embedded below.
'
BUILDING GRAPHS TO FIGURE OUT WHATS HAPPENING HERE
All graphs will be judged on worst residual Fstat. |Z score|< 3 is acceptable. Drift edges must be non zero. Some drift edges which show zero are actually non zero when decimals are looked at in the .ggg file.
On mobile, click on graph to see it in high resolution. On desktop, download the graph images and view in a zoomable picture viewer.
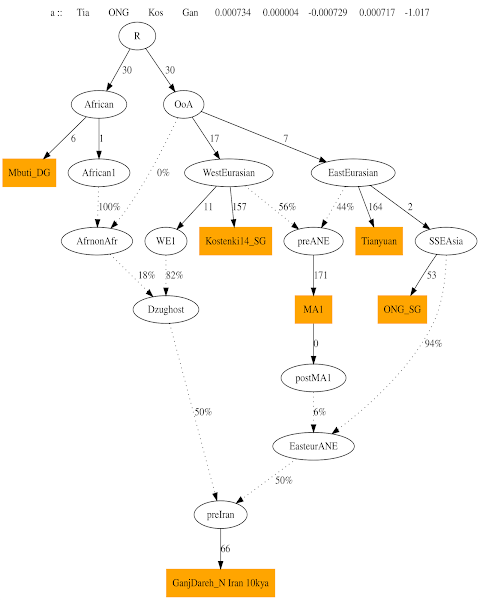
1. Basic graph with Mbuti, Kostenki, Tianyuan, Onge and MA1
2. Adding IranN
3. Adding CHG - Kotias
4. Adding EHG
5. Adding Irula (South Indian Tribe)
Adding Irula makes the IranN component split into two and thus we get the need for the node IndiaN. This node was discovered in the Rakhigarhi paper
2, but yet a name has not been given. So i have decided to call it IndiaN, although its formation is much prior to the neolithic, somewhere around 20-15 kya. Important to note is that IndiaN provides 40% of Irula ancestry, informing us of the deep impact of this ancestry even to the south of India.
WE NOW COME TO OUR TARGET POPULATION - STEPPE ENEOLITHIC
We will now test which EHG population and which Iran/CHG like population is chosen as sources for Steppe_En.
The options for the Iran like component are CHG, GanjDareh, IndiaN or commonIndoIran node.
1. I first tried the EHG + CHG model for Steppe Eneolithic.
The worst residual Fstat was
worst f-stat: ONG Gan Kot Ste -0.004638 -0.001959 0.002679 0.000494 5.418
The + Zscore told me that an additional affinity to GanjDareh was needed in the model.
2. Adding GanjDareh related node as a source
Worst Fstat still >+3. Tells me that SteppeEn still needs more affinity to Kotias (CHG). Which I will fix in next step.
3. Shifting CHG Source to postCHG from preCHG node.
It doesnt work, and the worst Fstat makes less sense. But these 2nd,3rd highest fstats need fixing first.
Tells us that there needs to be more affinity in the model between steppe_en and Irula. So in Next step we add IndiaN as source.
MA1 Iru ONG Ste -0.015150 -0.012315 0.002834 0.000628 4.510
MA1 Iru Gan Ste -0.010251 -0.007745 0.002506 0.000536 4.672
MA1 Iru Kot Ste -0.008653 -0.006722 0.001931 0.000641 3.014
MA1 Iru EHG Ste 0.001287 0.004926 0.003638 0.000652 5.576
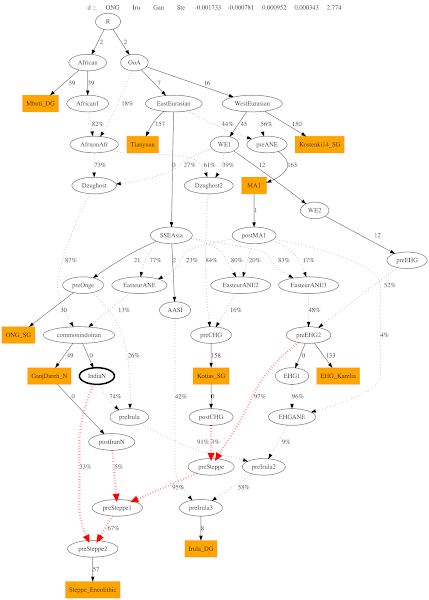
4. Adding IndiaN as source to Steppe_En
It works. Worst ZScore is < 3. The graph chooses IndiaN as a 33% source to steppe eneolithic, even with the option of IranN or CHG as sources. IranN and CHG shows only minor contribution.
DISCUSSION
I have shown evidence above that there is a very very high likelihood of an IndoIranian ancestry to have provided ancestry to both Indian groups like Irula (and IVC) as well as Steppe Eneolithic. My unpublished work shows that this same ancestry has provided significant ancestry to populations in SC Asia like Sarazm 3000bce. Where this source lived in 6th millenium bce is unknown yet, but in the vicinity of NW south asia and SC asia will be a good guess. Could be either early Mehrgarh culture or Jeitun culture but we need ancient samples from these cultures.
A similar conclusion was also drawn by Chad at populationgenomicsblog.com regarding the Steppe enolithic samples. His qpGraph work showed Steppe Eneolithic wanting south of caspian ancestry.
Recently, both Harvard and Max Planck seem to have changed their theory about PIE origins to Iran, and therefore the
post by Davidski on Eurogenes lamenting this. Harvard and Max Planck are of course on the right track, but it will be a while till their ideas move more eastwards towards South Asia and South Central Asia, but that will indeed happen.
References
1. Wang, CC., Reinhold, S., Kalmykov, A. et al. Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat Commun 10, 590 (2019). https://doi.org/10.1038/s41467-018-08220-8
2. Shinde V, Narasimhan VM, Rohland N, et al. An Ancient Harappan Genome Lacks Ancestry from Steppe Pastoralists or Iranian Farmers. Cell. 2019;179(3):729-735.e10. doi:10.1016/j.cell.2019.08.048
3. All qpgraph output files for all the graphs are in this folder, along with qpgraph parameter file.
Also Read:
Edit:
1. Added Satsurblia as CHG too, so that the label CHG is pseudo diploid.
2. Added drift edges after admixture nodes to appease perennial whiners.










18 comments:
Wow! nice qpGraph!
Beautiful post.
Absolutely wonderful, good job on this work.
Good work but do genetics researchers realize that when they speak of language spread they are relying on unproven, even fabricated concoctions by linguists over the last 200 years, For example, while everyone knows that PIE is a language "reconstructed" by linguists no one asks about "proto-Indo Iranian" - a completely non existent language with zero evidence, conjured up simply to make a connection between wherever linguists want to place PIE with India and Iran. That aside "Avestan" is also a "reconstructed" language. No Zoroastrian text mentions that name and the reconstruction was done from texts that were written 2000 or more years after the founder Zoroaster. The name itself was taken from the work of a 18th century Frenchman called Anquetil du Perron. The problem of science is the reliance on linguistics cross references, which though peer reviewed, have never been validated.
nope, genetic researchers just paste references to david anthonys work in their papers and move on with their life haha.
for example, in the latest horse paper which I made a post about, the authors just assumed dom2 horses were brought to indo iranian region in the 2nd mill bce from sintashta without studying a single horse remain from the region.
No opposition to this BS in peer review.
Great analysis. Here’s an older post from FrankN that looks more closely at the concerned regions . https://adnaera.com/2018/12/10/how-did-chg-get-into-steppe_emba-part-1-lgm-to-early-holocene/
The archeology i mean
yea, nice post that one.
Great post. Refreshing to see new insight on these matters.
Great Effort 👏
One of the clearest possible inference from this is that perhaps this is how R1a z645 reached the steppe so early. It’s southern association is supported by the presence of high diversity of R1a lineages even ancestral to z645 in India and Iran.
Thanks all. a new post is up responding to some criticisms of the anthrogenica cabal.
"It’s southern association is supported by the presence of high diversity of R1a lineages even ancestral to z645 in India and Iran"
Hey Jaydeep, Don't such R1a lineages which are ancestral to Z645 found outside india and iran too ? I can see few such samples at yfull here @ https://yfull.com/tree/R-M459/ .
Could you possibly define your concepts in the beginning? For example, are "African" and "WE1" replicators, meaning long strings of DNA code? Or are they taxonomical groups that have been formed based on something? When you present conclusions, that pre-ANE received 54% from West Eurasian and 46% from East Eurasian, do you mean percentages of this genetic string? I am trying to build a conceptual framework that would be applicable for the evolution of human genes and populations, and also to names, languages and archaeological technologies. A cross scientific approach is useful, but a linguist or archeologist will not understand your diagrams properly, unless you start by defining the concepts and measurements you are using. If you start with such basics, your article will also be more interesting for the wider audience.
If needed, I can help you find peer reviewers or a publisher.
Humans migrating from colder regions to Tropical regions makes more common sense than Humans moving from Tropical regions to Colder regions.
@pasi
The orange nodes are actual ancient dna samples, the white nodes are hypothetical populations.
The admixture nodes (and the %s) inform us about what % each of the 2 source populations provided to its target.
People may have moved north after the ice age thaw
Good research.
Post a Comment