ABSTRACT
qpGraph modeling of the Steppe_Eneolithic samples shows that they received up to 40% of ancestry from the common ancestors of the later Indus Valley Civilization (IVC or SSC). The Steppe_Eneolithic ancestry from the piedmont steppe from 4336-4047 BCE is the major source of subsequent cultures like Yamnaya & Corded Ware, which are widely suggested to be the vector of the language spread of European language families. This genetic connection, therefore, offers a significant genetic clue to the common ancestor of the first Proto-Indo-European language speakers.
THE SOURCE OF THE YAMNAYA ANCESTRY
The Yamnaya culture or culture with an autosomal ancestry profile similar to that of Yamnaya is widely believed to be the source of the Indo-European languages of Europe.
Western and Eastern Europe came into contact ∼4,500 years ago, as the Late Neolithic Corded Ware people from Germany traced ∼75% of their ancestry to the Yamnaya, documenting a massive migration into the heartland of Europe from its eastern periphery. This steppe ancestry persisted in all sampled central Europeans until at least ∼3,000 years ago, and is ubiquitous in present-day Europeans. These results provide support for a steppe origin of at least some of the Indo-European languages of Europe. - Haak et al, 2015 (1)
The source of the Iranian-like ancestry in Yamnaya has always been a mystery. The confusion flows from this formal F3 admixture test where both CHG (Caucasus Hunter-Gatherer, 2 samples found in Georgia) and IranN (Ganj Dareh Zagros herders, 10kya samples from Zagros mountains) along with EHG as source show negative F3 and significant Z-Scores, indicating that both are plausible sources.
Below is a Twitter conversation between Dr. Mathieson from U. Penn and Dr. Laziridis from Harvard University. It is quite clear that neither Caucasus Hunter-Gatherers (CHG) nor Iran Neolithic (IranN) samples from Zagros quite fits the bill by themselves as a second source of the Yamnaya ancestry.
Wang et al, 2019 (2) found 3 samples from the Piedmont Steppe north of the Caucasus mountains dated to 4336-4047 BCE which proved to be very good fits as the ancestors of the Yamnaya culture. However, even this study could not pinpoint the actual source of the CHG/Iran ancestry.
North of the Caucasus, Eneolithic and BA individuals from the Samara region (5200–4000 BCE) carry an equal mixture of EHG- and CHG/Iranian ancestry, so-called ‘steppe ancestry’ that eventually spread further west, where it contributed substantially to present-day Europeans, and east to the Altai region as well as to South Asia. - Wang et al, 2019
 |
| Location of the Steppe Eneolithic Samples at Progress & Vonyuchka |
The latest research. Chintalapati et al, 2022 preprint (13), shows that the admixture between EHG and Iranian-related ancestry in Yamnaya and Afanasievo occurred around 4400-4000 BCE.
To understand the timing of the formation of the early Steppe pastoralist-related groups, we applied DATES using pooled EHG and pooled Iranian Neolithic farmers. Focusing on the groups with the largest sample sizes, Yamnaya Samara (n=10) and Afanasievo (n=19), we inferred the admixture occurred between 40-45 generations before the individuals lived, translating to an admixture timing of ~4,100 BCE . We obtained qualitatively similar dates across four Yamnaya and one Afanasievo groups, consistent with the findings that these groups descend from a recent common ancestor.
Thus, we combined all early Steppe pastoralist individuals in one group to obtain a more precise estimate for the genetic formation of proto-Yamnaya of ~4,400 to 4,000 BCE.
These admixture dates give another line of evidence to the theory that Steppe_eneolithic were the ancestors of Yamnaya, and were the first in the region to possess this unique EHG + Iranian ancestry.
THE IndiaN ANCESTRY
Shinde et al, 2019 (3) concluded that the 'Iranian like' ancestry which gives between 60-87% ancestry to the Indus Periphery samples is not the same ancestry as that of IranHG or IranN, but rather they share a common ancestor deep in time (split before 10000BCE). Since the study does not name this ancestry, I have decided to name it IndiaN, which will be used in my subsequent models. The location of these IndiaN people prior to mixing with AASI people of the Indian subcontinent is unknown, but they could well be inhabitants of NW India since their split with IranN 12000 ya, or they could be more recent migrants to NW India around 7000 ya (5000 BCE) where they mixed with AASI. We just don't know yet due to a lack of relevant samples. The IndiaN ancestry is so named not because we know it's the original location, but because it provides the largest chunk of the ancestry in modern people of the Indian subcontinent today.
 |
| Fig3 from Shinde et al 2019 shows IndiaN split first from a common ancestor |
Using qpGraph(Patterson et al., 2012), we tested all possible simple trees relating the Iranian-related ancestry component of these groups, accounting for known admixtures (Anatolian farmer-related admixture into Hajji Firuz and Tepe Hissar and Andamanese hunter-gatherer-related admixture in the IVC Cline)(Figure S3), using an acceptance criterion for the model fitting that the maximum Z scores between observed and expected f-statistics was <3 or that the Akaike Information Criterion (AIC) was within 4 of the best-fit (Burnham and Anderson, 2004). The only consistently fitting models specified that the Iranian-related lineage contributing to the IVC Cline split from the Iranian-related lineages sampled from ancient genomes of the Iranian plateau before the latter separated from each other.
This qpGraph method (5) is what we will use to build our tree for Steppe_Eneolithic, which helps us overcome the limitation of the absence of IndiaN samples from the aDna record.
BUILDING THE TREE MODEL
The following sample labels from the available ancient and modern DNA datasets will be used to build the tree.
1. Mbuti.DG - Pygmy group from Modern Congo as Outgroup
2. China_Paleolithic - 2 samples, one from Tianyuan ~40kya and one from Amur River China ~33kya
3. Ong.SG - Modern Onge Andamanese
4. Yana_UP.SG - 2 samples from NE Siberia Yana RHS region, ~32kya, ancestry label ANE
5. Tarim_EMBA1 - 12 samples from Tarim Basin China, ~4-3.5kya, related to ANE ancestry
6. IronGates_Mesolithic - 29 samples from Serbia, ~9-8kya, also labeled WEHG
7. EHG_Karelia - 2 Samples from Western Russia, ~8.5kya, also labeled EHG
8. CaucasusHG or CHG - 2 samples from Georgia, ~13.5 - 9.5kya
9. GanjDareh_N - 10 IranN herder samples from Zagros, W Iran, ~10kya
10. IVC - 6 outlier samples from Shahr_i_Sokhta which are migrants from IVC, ~4.5kya
11. Steppe_Eneolithic - 3 samples from Piedmont steppe, ~6.4-6 kya
Model Parameters and Acceptance Rules
The common parameter file for qpGraph is pasted below. Allsnps: YES is used along with Inbreed: NO
1. Z-Score threshold is set at 3, bad fits will be reported if the model has F Stats with Z-Scores above +3 or below -3, and we will look for a better model.
2. All drift edges must be of non-zero drift length.
3. Sometimes, a 0 drift length edge as seen on the graph is actually non-zero (but minuscule) when the output file is inspected.
Starting from Scratch
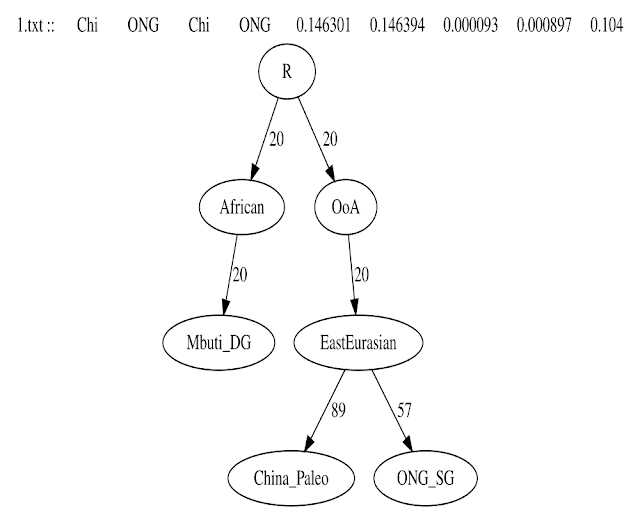
1. Adding Mbuti, China_Paleo & Onge
2. Adding Yana_UP
4. Adding IronGates_Mesolithic
6. Adding CHG_Georgia
7. Adding GanjDareh_N
8. Adding IVC
Some Notes on the findings of the preceding qpGraphs
1. Shahr_I_Sokhta IVC or Indus_Periphery is modeled as 29% AASI and 71% of IndiaN1 which itself is an admixture of IranN related ancestry (IndiaN) and 11% Tarim Basin related ancestry (related to ANE and WSHG). This is consistent with the conclusions of Shinde et al, 2019 & Narasimhan et al, 2019.
2. The hypothetical component AASI shares a common ancestor with Onge Andamanese deep in time, consistent with Narasimhan et al, 2019.
3. China Paleolithic and Onge share a common East Eurasian ancestor, corroborating with He et al (6), 2021. Minor archaic Human admixture in China_paleolithic is not seen as we havent included Denisovan or Neanderthal in our tree.
4. CHG and IranN don't form a clade, consistent with Laziridis, 2018 preprint (7). CHG and IranN obtain 60% and 50% ancestry from a West Eurasian source respectively, consistent with Laziridis 2018 preprint. We also detect ~8% and ~14% basal/African ancestry in CHG and IranN respectively, which is close to the estimate of Laziridis 2018 preprint. The remaining ancestry in our tree is East Eurasian, unlike Laziridis 2018 which gives a mixture of ANE (MA1 related) and east Eurasian. This could be because Laziridis 2018 used the actual Dzudzuana sample in their paper, whereas we use a proxy WE node as the Dzudzuana sample has not been published yet.
5. EHG is 40% West Eurasian + 60% ancestry related to ancestors of Tarim_EMBA1. This is close to Laziridis et al., 2016 (8) model of 25% West Eurasian + 75% ANE related to AG2 (or MA1) as Tarim_EMBA is more east Eurasian Shifted than MA1 or AG2.
6. Tarim_EMBA is modeled as 70% ANE (related to Yana and 30% East Eurasian (related to the deep ancestor of Onge). This is close to Zhang et al, 2021 (9) model of 72% AG3 + 28% Northeast Asian ancestry.
7. This is the basic tree that we will work on in our next step. Please note that this is not the only possible tree for fitting all the samples, and neither can we say that this is the best fitting model. However, it passes all our criteria and we can proceed with the analysis using this as a baseline. The worst fit F stat has a Z-Score of 2.03 <3, and all the drift edges are non-zero. The edges that have a 0 label on them are rounded off by the program, the actual non-zero values of which can be seen in the respective .ggg files (supplement).
FITTING STEPPE_ENEOLITHIC
Now that we have the base model setup, it is a matter of adding Steppe_eneolithic to the graph and seeing what sources give the best fits. We will test various models one by one - choosing between EHG and CHG/IranN/IndiaN as sources, as a 2 source, 3 source, and 4 source model.
1. Steppe = EHG + CHG
A bad fit with worst ZScore of 6.6 for f4 (Yana, IVC; CHG, Steppe_En) indicating that Steppe needs ancestry from a source close to IVC. There are total 91 outlier F stats, each signaling a need for either IVC or IranN ancestry.
2. Steppe = EHG + IndiaN1
Since the previous model showed the need for IVC-related ancestry, we will try this model.
The model fails with the worst Z Score of -10 for f4 (Irongates, Steppe; CHG, IVC), signaling that CHG ancestry is needed for steppe. There are 100+ outlier F4 Stat, most of which signal a need for CHG ancestry in the steppe.
The model fails with the worst F Stat Zscore = 11.15 for f4(Irongates, CHG; IraN, Steppe) signaling a need for CHG.
Outlier Z Scores
With the above (failed) models, what we see is that apart from EHG, at least 2 sources for Iranian components are needed in Steppe_EN, CHG & IndiaN.
4. Steppe = EHG + CHG + IranN
The model fails with worst F4 Z-Score of 4.64 for f4(Yana, IVC; IranN, Steppe) signaling that there is still a need for IVC-related ancestry. All the outlier F4 Z-scores also signals the same need. They are pasted below.
5. Steppe = EHG + CHG + IndiaN
Model is a success with the worst f4 Z-Score of 2.52. IranN ancestry is not needed, unlike in the above model where IVC-related ancestry was needed.
Steppe_Eneolithic = 50%EHG + 44% IndiaN1 + 6% CHG +- standard errors
FINAL MODEL
From the above, we can see that the that the only working model for Steppe_Eneolithic is a 3 source model of EHG + CHG + IndiaN with EHG and IndiaN being the major components.
However, the model whihc provides the best fit is a 4 source model which also includes IranN. It gives no F4 outliers, the lowest final score (best fit) as well as all non zero drift edges.
Worst F4 Z-Score 2.7. Steppe = 51% EHG + 40% IndiaN1 + 6% CHG + 3% IranN
DISCUSSION
No study so far has delved into the nature of the source of the Iranian related ancestry in the Steppe Eneolithic. The above qpGraph models conclude that the only models which fit for Steppe_en have to include EHG, CHG, and IndiaN as a source. The western Iranian Zagros herder-related ancestry has little part to play in the genesis of the Steppe ancestry. The graphs show a high prevalence (40%) of ancestry related to the ancestors of IVC people in the steppe profile. The usual caveats apply - we need to find the actual samples corresponding to the 5000-4000 BCE time period from the NW Indian subcontinent, SC Asian, and Eastern Iranian regions. qpGraph outputs may also change with the inclusion of other reference groups.
Narasimhan et al., 2019 provide the admixture date between AASI and IndiaN components in IVC samples from Shahr-i-Sokhta as 4483-3811 BCE. This could have occurred in two ways:
1. The IndiaN ancestry in IVC (node IndiaN3) resided near the IVC region and it was the AASI ancestry that moved to NW Indian subcontinent for the admixture.
OR
2. The AASI ancestry resided near the IVC region and IndiaN3 ancestry admixed with this ancestry from the west of it.
Ancient DNA from the NW Indian subcontinent region from 5000 BCE should give a definitive answer to this question. What is clear is that the same ancestors of IVC people gave ancestry to both IVC and Steppe post 6000 BCE, which provides evidence to explain the common source of Indo & European languages.
THE INDO IRANIAN BRANCH OF IE
The linguistic case for a steppe homeland (Sintashta/Andronovo Horizon) of Proto-Indo-Iranian language assumes a (one or more) non-IE language substrate in NW of the Indian subcontinent. The hypothesis assumes a complete replacement of extant languages by the incoming people (predominantly male) from the steppes. However, there are many inconsistencies with this hypothesis as has been laid out in a
comprehensive article by Jaydeepsinh Rathod (11), using tens of references from the work of linguists. The article also argues for a much older presence of the IE language family in the Indian subcontinent than is proposed by the steppe theory (1500BCE). There have been other historians, linguists, and archaeologists who have also argued for a much older presence of IE languages in the subcontinent. This paper agrees with the assertion of the older presence of IE languages in the NW Indian subcontinent given the genetic contact between ancestors of IVC and Steppe_eneolithic.
The answer to the question regarding the linguistic nature of contact between the bronze age Steppe people and Indo Iranian speaking people of the Indian subcontinent & Iran remains unclear because of the lack of any extant texts from the steppe. It is clear that the steppe bronze age ancestry appeared in the Indo Iranian speaking regions post-2000 BCE, but the nature of linguistic contact, loanword exchange, etc needs more study.
THE PROTO-INDO-EUROPEAN HOMELAND
The crux of the above analysis is that the same source that provides the maximum amount of ancestry to modern Indians (especially north Indians & Pakistanis), also provides a big chunk of ancestry to the Steppe component. This steppe component is believed to be the vector of language spread to the ancestors of most IE-speaking Europeans today.
This validates the theory of an Iranian PIE homeland, also supported by
Johannes Krause of Max Planck Institute in his 2021 book
(10). However, given that the bulk of the Iran-like ancestry in the Steppe is related to IVC ancestry rather than CHG or western Iranian herder ancestry,
the locus of PIE must be shifted to the east of what has been suggested in the book. Another reason why a northwest Iranian PIE is unlikely is that the region had a big amount of Anatolian farmer ancestry already by 6000 BCE, which is missing from the Steppe Eneolithic. The high in Anatolian Farmer ancestry 6000 BCE Hajji Firuz chalcolithic samples are evidence of this. The location of these samples is actually inside the locus proposed by Krause (in the graphic below)
This eastern locus supports the hypothesis by the anthropologist from St. Petersburg State University, Alexander Kozintsev (12) among others, who proposed east of Caspian sea origin of PIE.
 |
| From Chapter 6: A Short History of Humanity by J. Krause, Max Planck Institute |
THE PROPOSAL FOR PIE
 |
| THE BIG PICTURE: Genetic admixture events post 6000BCE can explain the IE Language dispersal |
TOOLS, DATA, AND OUTPUT FILES
1. The latest version of ADMIXTOOLS was used for qpGraph and convertf. Available
here.
2.
Plink 1.9 was used to make only a subset of required samples from the large eigenstrat database.
3. The genotype files and qpGraph parameter/input/output files are uploaded
here, which can be used for verification and rebuilding the models.
REFERENCES
1 Haak, W., Lazaridis, I., Patterson, N. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015).
https://doi.org/10.1038/nature14317
2 Wang, CC., Reinhold, S., Kalmykov, A. et al. Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat Commun 10, 590 (2019).
https://doi.org/10.1038/s41467-018-08220-8
3 Shinde V, Narasimhan VM, Rohland N, et al. An Ancient Harappan Genome Lacks Ancestry from Steppe Pastoralists or Iranian Farmers. Cell. 2019;179(3):729-735.e10.
doi:10.1016/j.cell.2019.08.048
4 Narasimhan VM, Patterson N, Moorjani P, et al. The formation of human populations in South and Central Asia. Science. 2019;365(6457):eaat7487.
doi:10.1126/science.aat7487
6 He G, Wang M, Zou X, et al. Peopling History of the Tibetan Plateau and Multiple Waves of Admixture of Tibetans Inferred From Both Ancient and Modern Genome-Wide Data. Front Genet. 2021;12:725243. Published 2021 Sep 3.
doi:10.3389/fgene.2021.725243
7 Paleolithic DNA from the Caucasus reveals core of West Eurasian ancestry
Iosif Lazaridis, Anna Belfer-Cohen, Swapan Mallick, Nick Patterson, Olivia Cheronet, Nadin Rohland, Guy Bar-Oz, Ofer Bar-Yosef, Nino Jakeli, Eliso Kvavadze, David Lordkipanidze, Zinovi Matzkevich, Tengiz Meshveliani, Brendan J. Culleton, Douglas J. Kennett, Ron Pinhasi, David Reich
8 Lazaridis, I., Nadel, D., Rollefson, G. et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016).
https://doi.org/10.1038/nature19310
10 Krause J, 2021 A Short History of Humanity- How Migration Made Us Who We Are Penguin Books. Chapter 6 Europeans Find a Language
13 Chintalapati M., Patterson N., Moorjani P. et al. Reconstructing the spatiotemporal patterns of admixture during the European Holocene using a novel genomic dating method
















70 comments:
First of all, I wanna say, absolutely great job! Your work has significantly advanced the current state of OIT studies (or Out of South Central Asia???). These days, people don't care about a theory at all unless they are provided genetic evidence. Hopefully this will allow scholars to take our theories seriously.
I notice you have written that you are unsure about the temporal and spatial placement of the India/IranN ancestry in India. This paper may have evidence of a NW Indian location of this ancestry: https://www.nature.com/articles/s41467-019-09209-7.
This paper basically says that early Anatolian farmers show a shift in affinity to IranN populations and also South Asian populations. See Figure 2. Jaydeepsinh writes "Coming back to the Anatolian Hunter Gatherer (AHG) paper, referring to figure 2a of the paper we can see that the Anatolian Aceramic Farmer (AAF) who is formed by the admixture of AHG : Iran_N in the ratio 90 : 10, shows the most ancestry sharing relative to AHG, with CHG and modern South Asians, among whom Mala & Vishwabrahmin top. Now, as we noted earlier Mala & Vishwabrahmin are heavy AASI groups. Yet, an admixture of Iran_N with AHG results in AAF who show the greatest shift towards AASI heavy populations from South Asia.".
Does this provide evidence for the placement of the IndoIranN ancestry next to AASI? Maybe the heavy intermixing picked up later during chalcolithic and bronze age. This paper also supports a south asian origin of farming. IndoIranN ancestry penetrates Anatolia during late pleistocene. Premendra Priyadarshi has written on the evidence showing farming coming from south asia.
I also want to note how Kozintsev supports a SC Asian homeland of PIE, agreeing with Nichols, despite the current state of Steppe dogmatism. Its refreshing to see some alternative work. I am looking forward to reading "Proto-Indo-Europeans: The Prologue". I have read his "On the Homelands of Indo-European and Eurasiatic: Geographic Aspects of a Lexicostatistical Classification (2020)", which was pretty interesting.
Thanks. I will read the paper you quoted, but I don't find the question very important now. I'm happy to wait for samples from the region for that time period.
I think the biggest obstacle for the steppe theory promoted by Harvard for india/iran is that it NEEDS a complete domination scenario to wipe out all traces of old languages from the region, by males, something which the swat dna hasn't been able to prove.. the indians still had 75-85% local ancestry..
Jaydeep's recent article makes short of the steppe theory's linguistic claims given this genetic backdrop.
New preprint just dropped today
https://www.biorxiv.org/content/10.1101/2022.01.18.476710v1
The EHG IranN admixture date in Yamnaya, Afanasievo has been dated to 4400-4100 BCE, which should also be the date of the formation of the steppe eneolithic.
" Our analysis reveals the precise timing of the genetic formation of these early Steppe pastoralists groups–Yamnaya and Afanasievo–occurred ~4,400-4,000 BCE. "
I don't know ABC of genetics but thought this might interest you all https://eurogenes.blogspot.com/2017/07/the-out-of-india-theory-oit-challenge.html?m=1
"whom Mala & Vishwabrahmin top. Now, as we noted earlier Mala & Vishwabrahmin are heavy AASI groups."
Wouldn't this mean that the iran_N like ancestry in these Anatolian aceramic farmers from 7k-6k BCE was already mixed with AASI and thus, populations like Mala and vishwabrahmin are giving stronger values in these D tests ?
"This paper also supports a south asian origin of farming." --- So, south asians taught anatolians how to grow wheat :) ?
> Wouldn't this mean that the iran_N like ancestry in these Anatolian aceramic farmers from 7k-6k BCE was already mixed with AASI and thus, populations like Mala and vishwabrahmin are giving stronger values in these D tests ?
yes, in small amounts, cause qpadm did not pick it up.
> "This paper also supports a south asian origin of farming." --- So, south asians taught anatolians how to grow wheat :) ?
Not wheat, which I believe is native to west eurasia. But there is evidence of introduction of some agricultural packages.
Wouldn't this mean that the iran_N like ancestry in these Anatolian aceramic farmers from 7k-6k BCE was already mixed with AASI and thus, populations like Mala and vishwabrahmin are giving stronger values in these D tests ?
It is very difficult to differentiate between east eurasian ancestries without actual AASI sample. east and west eurasian ancestries are pervasive throughout eurasia.
"This paper also supports a south asian origin of farming." --- So, south asians taught anatolians how to grow wheat :) ?
almost no chance of that, genetically speaking. The IranN ancestry present in anatolian farmers is very old and just from western iran.
"I don't know ABC of genetics but thought this might interest you "
Davidski, is that you. Sly bugger. Hahaha
Your are doing high level word play without more thorough investigation of sources of H1, L1, R1, R2, J2b2, J2a etc...
@vAsiSTha
Take a look at this recent paper, in Antiquity journal, showing links between Western Iran, Irak, and Maikop culture in northern Caucasus. Authors claim golden and silver drinking straws in Maikop are related to the same use in Mesopotamia.
https://tinyurl.com/vh9zszz8
Thanks carlos
@suresh
Falsify my finding through openly available tools that ivc and steppe_eneolithic don't share a common ancestor..
I feel that you are somewhat triggered by the use of 'IndiaN' for the ancestors of IVC.
It is a fact that this ancestry does not occur in significant amount in any population starting from bronze age itself apart from steppe and South asia (even today).
Already by bronze age we see Eastern Iran (shahr sokhta) and bmac deriving their ancestry from the zagros IranN (evidenced by high Anatolian component).
So the name chosen as IndiaN is purely because of its abundance in south asia today.
As far as Y haplogroups are concerned, let us first find 5000 bce samples from east iran, SC asia before the impact of west asian ancestry. Let's see what we get there.
@Vasistha
Which modern population (world-wide) continues to possess the highest percentages of Iran_Neolithic or India_N autosomal components?
And...of course, Which modern Indian population group displays the highest?
@bruin
Zagros ancestry is not found anywhere without Anatolian component. The admixture happened close to 6000bce itself (west asia being close to Anatolia).
This type of ancestry (mixed Anatolian, zagros) will likely be seen in the Mediterranean, and the caucasus but I will have to revisit these region samples to confirm.
This IndiaN ancestry is only seen in IVC samples and some in sarazm eneolithic (being eastern Turan).. all the western turanian samples (geoksyur, namazga, parkhai, shahr-i-sokhta, bmac) already by 3000bce show a big Anatolian component, so their iran ancestry is mainly zagrossian. But maybe minor local IndiaN sort of ancestry remains. We need the IndiaN and AASI sample to dig deep.
@vAsiSTha
How can you solve the problem to your analysis that there are some aDNA samples of R1a in Russia and Ukraine from around 10,000 to 8,000 BCE, and you consider IndiaN1 arrived in the steppes only around 6,000 to 4,000 BCE?
Please see:
https://tinyurl.com/xcrv4j5s
@carlos
From a new preprint that date 6000-4000 is now 4500-4000 BCE. This is when IndiaN1 and EHG mixed to form proto Yamnaya.
The R1a question remains open. With the data so far it seems there was a minor trickle of R1a from Europe before 2000bce, but that L657 mutated in 1 person in India itself (given the absence of L657 in steppe ancients or in modern Europe). L657 is an Indian and arab marker. There is 0 evidence of large number of L657 makes from steppe invading or migrating into SA.
As far as R1a map is concerned, Davidski's map is incomplete.
There is R1a also in maykop, Baikal eneolithic (lokomotiv) as well as Xioahe(Tarim)
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2838831/ - Xiaohe R1a
2 samples from Lokomotiv are listed as R1a also
LOK_1980.006 - Lokomotiv cemetery, Irkutsk R-M198 (R1a) F 7250 Russia - Lokomotiv_EN
LOK_1981.024.01 - Lokomotiv cemetery, Irkutsk R-M198 (R1a) F 7250 Russia - Lokomotiv_EN
Plus there's SA6013 from Maykop who is R1a1(xR1a1a).
@vAsiSTha
I can agree with you that L657 formed in India, but to me it´s more plausible that it corresponds to an early arrival of R1a-Y3 in India around 2200 BCE through Central Asia.
@carlos
Yes thats possible
Although even y3 is not found in any of the steppe sites.
First one is from alai nura 300ce, when Buddhist contacts from SA to China are well attested.
I thought Y3 was the version of Z93 carried by bronze age steppe tribes. In that case whhich version did Sintashta and Andronovo have?
Sintashta and andronovo have Z2124.
Z94 has 3 son clades -
Y3 and Y40 found mainly in SA and middle east. None among steppe ancient
Z2124 found in central asia. Many among Steppe ancients.
Z93(xZ94) is found in Fatyanovo 2600BCE.
It's an interesting model, I am skeptical of some aspects, but if I may give some feedback,
would you consider using an ancient East Asian in your analyses? Wouldn't such a sample help clarify the source of ENA in ANE-impacted populations?
I am surprised about the SEA-like so far north, so I am wondering if it's some type of non-Tianyuan East Eurasian/East Asian instead. Tarim was not part Onge but rather part Baikal_EN in the paper the samples were reported in. I believe it to be the same ENA population affecting all of West Eurasia, going by your graph. And I suppose MA1 would also show this non-Tianyuan ENA right?
Also, why label Basal as African? It seems to branch post OOA no?
@gamerjZ
yeah, i cant add too many populations in order to keep the graph as simple as possible. So I add 2 E Eurasians (onge and China_paleo), 2 ANE (yana & tarim), 1 west Eurasian related (Iron Gates) to ground the various related nodes of the graph.
The others are helpful in answering the question which I set out to answer about sources of steppe_en. (chg, iranN, IVC)
About ANE, it seems that all the ANE in the Holocene populations is rather related to the Tarim basin than Yana.
Tarim is not shown to be part Onge here, but its rather drifted (81 drift units between SEA1 and SEA2). The SEA2 may just be closer to Baikal-EN if I add that pop too, who knows. In which case node nomenclature will also change.
Basal branch works both from African node, or OoA node in some cases.
@Vasistha
I see what you mean, too many populations would likely confuse things.
"About ANE, it seems that all the ANE in the Holocene populations is rather related to the Tarim basin than Yana."
Yeah, that makes sense because they all seem to have ENA affinity on top of what ANE has. Yang et al (2017) had mentioned this and also Reich and Lipson (2017) as well as the Lazaridis pre-print.
"Tarim is not shown to be part Onge here, but its rather drifted (81 drift units between SEA1 and SEA2)."
Yes you are right, for some reason I missed that. Might hint at the southern origins model for East Asians, some papers have them as mostly Onge, minority Tianyuan (though it's more like Onge-related).
"Basal branch works both from African node, or OoA node in some cases."
From what I've noticed most West Eurasians seem symmetrically related to Africans when compared to Eastern Eurasians except if they have substantial Levant_N-related ancestry. I also think D(Chimp,African;CHG,WHG) is not significant but will have to check again. Imo, African ancestry enters West Eurasia with Natufians and related pops which probably don't give much to CHG/Iran_N etc. After all most Central and South Asians lack subclades of E which is associated with this gene flow event.
I forgot to mention it yesterday but I am intrigued at the unique ENA population admixing into Iron_Gates_HG. I wonder if Bacho Kiro or Ustishim have a role to play here given their locations, because anything from the East would in my mind affect the others first (EHG,Iran_N,CHG and so on).
Yes the extra EE needed specifically for IronGates struck me as odd too. I was trying to fit both EHG and IG as some ANE + WE, but somehow it was not working out for Irongates.
Very interesting and refreshing post. Where do you think R-Z2103 fits into all this?
I'll have to study R1b-Z2103 a bit more.
Did a mega drought topple empires 4,200 years ago?
https://www.nature.com/articles/d41586-022-00157-9
According to Scroxton, people abandoned the northern cities between 4,200 and 3,900 years BP, and the southern cities more gradually between 3,900 and 3,300 years BP — making the Harappan civilization a largely rural society.
Hope this adds into your research.
Your well wisher ��
Get well soon
From the literate cultures that we have decipheres, empirede were toppled, but with a succession. The Sumerians were replaced by Akkadians and Guttians. There was the first Intermediate period in Egypt. Around 1900, the Old Babylonian period began and the Middle Kindom (around 2000BC).
The abrupt change of Indus is mostly confined with the areas affected desiccation of the Saraswati. In the denser areas like the Doab, Punjab and the Gurati there was continuity. Perhaps not as many large urban areas, and we have been observing, there was no abandonment or migration, but a restructuring of society. A good analogy would be the West Roman empire. Instead of the Senate of Rome commanding the lands, with the godlike figure of the Emperor of Rome, there was instead the Church of Rome as the center of power, legitimizing many small kings.
The power was strong as the latin language spread within all domains of the Church of Rome. The process stopped in England, where French almost became the 1st language, helped by the Protestant Reform and with the Germans, where the grip was always too weak.
"yes, in small amounts, cause qpadm did not pick it up."
@3rdacc, Well, then the D stats should have shown closeness to a less AASI rich and more IndoIranN population than Mala, vishwabrahmin as the hypothetical IndoIranN should have had 0 AASI when it admixed with Anatolian Aceramic Farmers who are dated to ~8000 BCE. May be there are some issues with interpretation of D tests.
"Tarim_EMBA is modeled as 70% ANE (related to Yana and 30% East Eurasian (related to the deep ancestor of Onge)."
@Vasistha, I noticed this in your qpGraphs, that is, the vector of east eurasian ancestry in Tarim_EMBA is more closely related to SEA(South-east asian) ancestry than China paleolithic. Afaik, in the paper, the Tarim was modelled as ~72% ANE + 28% Baikal_EN. Since I am not familiar with East ancient aDNA, was there some migration from SEA to NE China region around neolithic ? If not, i wonder why the EE ancestry to Tarim_EMBA seems closer to SEA node than China_paleolithic node ?
Celts were people of Danu
Aryas were people of Saraswati
Are there other main Indo-European branches attached to river goddesses? Where does it stand in relation to CHG female mediated transfer of languages into the West?
If true?
"Ancient Greeks had three words for father: patér (father), táta (daddy) and páppa (daddy)."
Where did táta and páppa originate?
táta sounds similar to dada (grandfather)
Hey vasistha,
It seems that the date of 4400-4100 BCE for the supposed formation of Steppe_eneolithic like profile(a mix of EHG + CHG/Iran_N) may not be accurate. One user altvred applied DATES algorithm on Steppe_eneolithic and found out that the mixing between these ancestries happened nearly ~2800 years before the dates of these eneolithic samples i.e., around and earlier than ~7000 BCE.
https://musaeumscythia.blogspot.com/2022/01/when-did-western-steppe-herder-genetic.html
Steppe_eneolithic samples are different from the Yamnaya as Yamnaya samples have some EEF ancestry which Steppe_En samples have not. So, is it possible that the preprint's 4400-4100 BCE date is capturing the time of admixture between the EEF ancestry in Yamnaya rather than CHG/Iran_N and EHG.
I guess the multiple admixture events confound the dates of admixture as mentioned in Narasimhan's paper
(Interesting the Nick Patterson has commented on the above post too).
"EE ancestry to Tarim_EMBA seems closer to SEA node than China_paleolithic node ?"
China paleolithic seems to be sort of a dead end. both east asian and onge seem to be closer to the SEA node, than china_paleo. however, the EE in tarim is quite drifted from Onge.
"One user altvred applied DATES algorithm on Steppe_eneolithic and found out that the mixing between these ancestries happened nearly ~2800 years before the dates of these eneolithic samples"
I trust the preprint better as it clumped a lot of samples together for EHG and lot of Iran related samples together for Iran as the 2 source pools.
The paper itself says that this is the admixture time between the EHG pool and IranN pool.
But generally, my trust in accuracy of DATES is now reduced.
"Did a mega drought topple empires 4,200 years ago?
https://www.nature.com/articles/d41586-022-00157-9
According to Scroxton, people abandoned the northern cities between 4,200 and 3,900 years BP, and the southern cities more gradually between 3,900 and 3,300 years BP — making the Harappan civilization a largely rural society.
"
Thanks
Could Rig Vedic Vritra be this 4.2kbp event? Indra helped release cosmic waters that lead to the revival of rivers?
possible, vedic ppl seemed to be quite perturbed by blockage of rivers and invoked many of the Gods to help them ease the water stress.
RV VI.61 has a mighty Sarasvati, but at the end it says: "O Sarasvati, lead us to a better state. Do not spring away with your milk; do not come up short for us. Take delight in our partnerships and communities. Let us not go from you to alien dwelling places."
The drying of the saraswati is also recorded. This is a 2300-1800bce phenomenon.
My comment on Davidskis latest post about PIE. It is not some curious artifact of steppe_En that it shows affinity to IVC. This affinity can be clearly seen in a basic qpAdm tests. The generated D-stats (significant ones above 3) tell us why the ehg+chg model fails with respect to actual samples. A clear consistent pattern is dearth of IVC related ancestry wrt multiple other reference populations. This is why an input from ancestor of IVC is required in the above qpGraphs.
"Some things have to be made very clear:
1. For the ancestors of Yamnaya, There is no purer ancestry found than Steppe_en at progress and vonyuchka. Tt is 50-50 EHG - CHG/Iran. There is no WHG component at all. There may be minor traces of Anatolian.
Any ancestry found north of Caucasus with steppe_en profile but also some WHG, will be considered an intermediate population to Yamnaya. Davidski will claim that this intermediate population is some new exotic stuff with no relation to Iran but that does not make it so.
so unless a population is sampled that has a higher CHGIran:EHG ratio than even steppe_en, steppe_en will remain by default the original source of Yamnaya.
2. The confusion between CHG and IranN comes because f3(Steppe_en; EHG,CHG) as well as f3(Steppe_en; EHG, IranN) are both significantly negative.
There are 2 populations to the south of caucasus range from the same timeperiod - areni_c, seh_gabi and meshoko. all have significant anatolian ancestry, the kind absent from steppe_en.
3. In the G25 models of areni_C, meshoko and Steppe_en; why does only Steppe_en pick up shared drift with Sarazm?
4. In qpGraphs, why does CHG+EHG or IranN+EHG fail due to affinity for an eastern population?
5. in qpAdm of steppe_en as EHG + CHG, why does the model fail due to d-stats showing less affinity with IVC in model than actual? https://pastebin.com/kv3CQY6K
EHG + CHG. p-val=4.2e-15
gendstat: China_AmurRiver_LPaleolithic IVC -3.438
gendstat: Yana_UP.SG IVC -5.262
gendstat: Serbia_IronGates_Mesolithic IVC -4.343
gendstat: ONG.SG IVC -4.518
gendstat: Mbuti.DG IVC -5.557
I do not expect an honest answer from Davidski"
"I trust the preprint better as it clumped a lot of samples together for EHG and lot of Iran related samples together for Iran as the 2 source pools.
The paper itself says that this is the admixture time between the EHG pool and IranN pool.
But generally, my trust in accuracy of DATES is now reduced."
I understand but considering Yamnaya had some ANF like ancestry which Steppe_En groups didn't, wouldn't that confound the admixture dates when one applies DATES ? I have been wondering but why didn't they apply DATES on the Steppe_En samples themselves just to cross-verify their results ? Anyways, it's a pre-print and who knows, may be the final peer-reviewed version will get changes .
Because the number of steppe_en samples are too small I guess. Dates averages out the per-sample covariance coefficient.
>
Could Rig Vedic Vritra be this 4.2kbp event? Indra helped release cosmic waters that lead to the revival of rivers?
Way too late. Rig Veda is pre IVC, as shown by Kazanas and Priyadarshi. Atharvaveda is contemporary to IVC.
That verse doesn't seem to indicate drying but awareness of droughts.
Any idea what kind of metals and weapons they used?
I am reading Mandala 6 (english) and it appears, bow & arrow to be the primary choice of weapon and one reference to stone weapon and Iron (100% sure it mis-stranslation).
Rig Veda 6.16.47-48
Agni, we bring thee, with our hymn, oblation fashioned in the heart.
Let these be oxen unto thee, let these be bulls and kine to thee.
The Gods enkindle Agni, best slayer of Vṛtra, first in rank,
The Mighty, One who brings us wealth and crushes down the Rākṣasas.
@AshishKaull
>I am reading Mandala 6 (english) and it appears, bow & arrow to be the primary choice of weapon and one reference to stone weapon and Iron (100% sure it mis-stranslation).
I am reading Kd Sethna rn, and apparently there are multiple translations of "ayas" by different scholars. I will look into this more, but I highly doubt there is iron. Even Witzel says this.
Yes that is what I have heard.
"Ayas" is that copper or bronze?
I don't know yet. Based on my dating of the RigVeda, ayas can not be any metal at all. But I have to look more into it.
Ayas without any adjective is just metal. Presumably copper or bronze.
Proto-Indo-European
Alternative reconstructions
*áyos[1]
Noun
*h₂éyos n[2][3][4]
a metal, copper, bronze
Usage notes
This is the only word in Proto-Indo-European that unequivocally refers to a metal. There is no word for iron and the words for gold and silver seem to mean “that which shines”, or “the golden” and “the silvery”, respectively. In the early Indo-European languages, this word refers to copper (and bronze), and the Proto-Indo-European word refers with absolute certainty to one of these metals, or both. This shows that the Indo-European language was spoken during a time when copper was used.
https://en.wiktionary.org/wiki/Reconstruction:Proto-Indo-European/h%E2%82%82%C3%A9yos
Thanks 🙏
"China paleolithic seems to be sort of a dead end. both east asian and onge seem to be closer to the SEA node, than china_paleo. however, the EE in tarim is quite drifted from Onge."
Just checking up on this post but I think Tianyuan being more of a dead-end may be the reason why East Asians appear closer to SEA node rather than actual Onge ancestry. Wang et al (2020) had East Asians sharing more drift with Onge than Tianyuan from what I recall, which is probably what this graph is showing although this ancestry may have been in northern Asia as far back as 30kya.
PS: @Vasistha have you seen these papers? https://www.biorxiv.org/content/10.1101/2021.11.04.466891v1 (about genetic continuity in Central Asia since the Iron Age) and https://www.biorxiv.org/content/10.1101/2022.01.31.478487v1 (about Neolithic Upper Mesopotamia) Y
I am linking the 2nd one, mostly because they report one sample carrying YDNA C2e1a1a which if not a fluke (high chance of that too imo)etc, and since all samples have Iran_N ancestry may hint at possible influences from NE Asia in Iran_N? C2e seems most common in Koreans and Japanese
@AshishKaull
"Papa" "Mama" sound similar in many languages, so although obviously there is some connection between IE languages in India and Europe, that's probably not it
@gamerz_j
I have two questions
1) What, in your opinion, is the origin of Y DNA K2b? It is found in Tianyuan who appears to be basal EA, can we trace the K2b from an EA male ancestor into western pops?
2) How about K2a which appears in western Eurasia pops like Ust-Ishim and Oase 2 but completely absent in said regions and found mostly in Eastern Eurasia (N-M231 and O-M175)?
@Ganesh
1) About K2b yes it most likely arrived in the west from an eastern pop, perhaps in the north or Central Asia someplace since Yana (31kya) already shows P* (or P1* can't recall which one rn).
2)K2a like K2b both most likely arose in an IUP-related population of which Ust-ishim, Tianyuan, Oase2 were all members of, but some with more archaic ancestry than others. Since East Eurasians are mostly or entirely of IUP descent it makes sense both lineages survive more in East Eurasia.
That's my opinion at least, don't know anything more specific than that.
The central asia paper didnt have many new samples of relevance. during the post buddhist period, the data confirms SAsian ancestry travelling on route to China.
Im reading the neolithic Mesopotamian paper now.
@Vasistha
Since you mentioned it I wonder how far north does SAsian ancestry extent in Asia? I think it's present (in low amounts) all the way in Xinjiang but different papers show different results.
Lots of common Y Hgs between china and SAsia including. We know that buddhism reached the tarim basim from SAsia (and hindu/buddhist deities all the way to Japan).
Unrelated to the topic at hand but have you seen this paper?
https://linkinghub.elsevier.com/retrieve/pii/S0378111921006934
Sharbadeb Kundu et al (2022) "The impact of prehistoric human dispersals on the presence of tobacco-related oral cancer in Northeast India"
They make some interesting arguments about when and how certain variants arrived in India.
Their conclusion was that NE India/South Asia was settled from the east, initially around 54kya and then there was another SEA-originated migration about 40kya. I am skeptical that's the case but perhaps they are hinting at a northern route for East Eurasians? I suspect their results may be biased by the East Asian ancestry of NE Indian populations.
@gamerj_z
Haven't read it yet, will read.
New post is up
Please don't get me wrong, it's just a hypothetical question*:
Would it be easy for genetic institutions to plausibly fake ancient DNA/RNA samples? As far as I know, simulacra are widely used already. Is that the case? Would they theoretically possess the means? Would they be indistinguishable from real samples?
*(irrespective of elaborating on (im-/)possible motives)
@eco
Yes its easily possible.
Can admix 2 samples, can add ancient dna random damage at ends of the strands.
After all it's digital data, can manipulate it at will i guess.
Thanks vAsiSTha.
//The impermeability of Anatolia to exogenous migration contrasts with our finding that the Yamnaya had two distinct gene flows, both from West Asia, suggesting that the Indo-Anatolian language family originated in the eastern wing of the Southern Arc and that the steppe served only as a secondary staging area of Indo-European language dispersal.//
https://iias.huji.ac.il/event/david-reich-lecture
this is huge!
Post a Comment