In my previous post, I analyzed the so-called 'Indus Periphery' samples from the data published in Narasimhan et al 2019.
My conclusion was that the IVCp samples can be best modeled distally as
Ganj_Dareh + Ancient North Eurasian (Tarim_Basin or West Siberian HG) + Onge + Levant_PPN or Anatolian Farmer.
In this post, I shall do an in depth analysis of the genome of the lone Rakhigarhi sample (Id: I6113, female) dated to around 2000 BCE, published in Shinde et al 2019.
 |
| Picture of I6113 burial, Rakhigarhi. From Shinde et al 2019 |
They could only extract 31.7k good quality SNPs from the aDna of this sample out of 1240k, so we are working with low quality data here. Nevertheless, this is an opportunity to find out the precision and power of the tools we use to detect admixture percentages.
SETTING UP qpAdm
We will model Rakhigarhi distally using a 2 source rotating model first. 2 sources from the following reference populations will be tested one by one as sources (except Mbuti/Central African) with the other populations as reference. A p-value>0.05 will pass the model, a p-value > 0.0001 will make the model suitable for further tests.
References populations used:
Mbuti.DG, CHG, EHG, China_YR_MN, Russia_Shamanka_En.SG, WSHG, PPN, Serbia_IronGates_Meso (WHG), ONG.SG, Iran_GanjDareh_N, Tarim_EMBA1, Turkey_N, Russia_MLBA_Sintashta
Settings: ALLSNPs set to 'YES'. Inbreed set to 'NO'.
2 Source qpAdm Models: P-Value Matrix
We can see from the above matrix that the only sources which come close to giving us a good solution
are (IranN or CHG)+ (Onge or Shamanka_En).
This is already an incredible amount of elimination that we have been able to do, in spite of the low quality sample. But we see that no 2 source model has crossed our p-value threshold of 0.05. So in the next step, I add a 3rd source one by one to our 4 best 2 source-models.
3 Source qpAdm models: P-Value matrix
At this moment it is worthwhile to note that Shinde et al proposes 2 models for Rakhigarhi, both of which are 2 source models viz. IranN + Onge, and Iran + Shamanka.
Already with the above analysis, we see that the more standard 3 source model as outlined in Narasimhan et al is preferred. The 2 source models for Rakhigarhi can be rejected.
From the above table, CHG + Onge + IranN has a good p-value of 0.165, but the proportions are all wrong with IranN scoring above 100% and CHG score -27%. So those models can be rejected as well.
So, CHG + ONG group has 1 acceptable model; CHG + Shamanka group has 1 as well, IranN + Onge group has 3 acceptable models, and iranN + Shamanka group has 1. Due to the low quality of the sample we can see that qpAdm doesnt have enough power to precisely eliminate the last few standing models.
Because, IranN + Onge group has the most number of acceptable models, and because of knowledge about the ancestry composition of IVCp samples, we choose these as the models for Rakhigarhi at the end of this step.
a) IranN + Onge + EHG (61%, 30%, 9% with Std Error 5.4%, 4.4%, 3.7%)
b) IranN + Onge + WSHG (64%, 29%, 7% with Std Error 5%, 4.8%, 3.5%)
c) IranN + Onge + Tarim_EMBA1 (65%, 27%, 8% with Std Error 4.8%, 4.7%, 3.9%)
EHG as a source cannot be rejected due to low quality of the sample and EHG sharing many alleles with WSHG/Tarim. As EHG can also be roughly modeled as WSHG + Western Hunter Gatherer (Serbia_IronGates) forming the Siberian-European cline.
We can already see here that our model for Rakhigarhi is converging towards that for IVCp, including the admixture coefficients. It is looking increasingly likely that the IVCp samples are representative of individuals like that from Rakhigarhi. So in my final step, I tried the rotating model on the IVCp samples to eliminate the last few models.
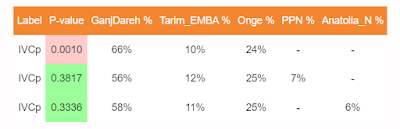
3 Source qpAdm models for IVCp: P-value Matrix
For IVCp, only 1 model viz. IranN + Onge + Tarim_Emba passes the P>0.0001 threshold. But it still does not cross p>0.05, so we test with a 4th source.
The only model which passes with p>0.05 for IVCp is
IranN + Onge + Tarim_EMBA1 + Turkey_N
(58%, 24%, 11%, 7%, Std Error 1.1%, 2.7%, 0.9%, 1.8%)
Importantly, we can also reject Sintashta_MLBA related steppe ancestry in IVCp as the models fail.
FINAL STEP: ELIMINATING THE LAST FEW MODELS FOR RAKHIGARHI
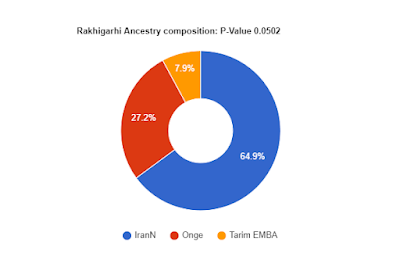
From the list of 3 possible models for Rakhigarhi, we can see that the only one which matches IVCp is model c) IranN + Onge + Tarim_EMBA1.
Although, Turkey_N is not needed to model Rakhigarhi successfully, unlike for IVCp, this may just be due to the low power. qpAdm is known not to detect need for minor admixture for low quality targets. We would just have to wait for better bronze age samples from Indian contexts to answer this question with certainty.
However, I went ahead and modeled Rakhigarhi with a 4th source Turkey_N.
The standard errors are large, and at 95% confidence (2 x SE), the anatolian component in Rakhigarhi could range from 3.3% to 36%. There is no reason to assume a large number, and if at all Anatolian ancestry in Rakhigarhi is to be assumed, a low 3-10% makes most sense given that we know IVCp composition.
CONCLUSIONS & CAVEATS
My conclusion differs from that of Shinde et al and Narasimhan et al on 3 points
1) I reject 2 source models for Rakhigarhi as concluded by Shinde et al, and propose that a 3 source model is must. That 3rd source is related to Tarim_EMBA1.
2) I reject WSHG as the 3rd source for IVCp as concluded by Narasimhan et al, and find that Tarim_EMBA is a better source compared to WSHG. Tarim samples were only published in 2021 so the authors of Narasimhan et al 2019 did not include them.
3) As outlined in my previous post, I do not accept the conclusion of Narasimhan et al 2019 that anatolian ancestry is absent in the IVCp samples. Rather, I find that it is necessary to model IVCp. For Rakhigarhi, Anatolian ancestry is not necessary, but high quality data from same time period is needed to confirm.
As a general rule for qpAdm, a source which passes in the model does not mean that is the true source, rather it means that a related population is the true source. Example, Onge admixture does not mean that the Andamanese admixed in Haryana, rather it means that a population which shares ancestors with Onge mixed in Haryana, and Onge is the best proxy we have so far.
All qpAdm results in drive folder
Update:
Ran 495 rotating qpAdm models on IVCp, 12 candidates, 4 sources at a time. I made a new python script to collate all the results in one file.
Only 1 model out of 495, which we have discussed above, passes.







22 comments:
I have modeled all the SC asian eneolithic samples and none of them require EHG ancestry, check previous post (even with ehg in the outgroup). The affinity you see is because of CHG like presence.
Sarazm especially doesn't have any steppe related ancestry.
Let me Rebut this Anthrogenica user's argument properly.
1. Sarazm samples are dated and calibrated to 3600bce. CentralSteppe_EMBA samples like Kumsay_EBA are dated to 3100bce. (all 4 kumsay samples are related, 3 dated to 3100-2900bce, one being 3345-3100bce. This outlier is also closely related, so likely that his date is closer to 3100bce).
The 3 Mereke (also steppe_emba) samples have a wide range between 3300 & 2000 bce. So we have established, Sarazm samples to be older by 300-500 yrs.
2. Sarazm needs no EHG as source in qpAdm modeling. Models work when EHG and Khvalynsk are in reference pops, therefore denying any direct steppe_en or steppe_emba impact on Sarazm.
Tajikistan_C_Sarazm
Iran_GanjDareh_N: 53% +- 5.8%
Turkey_N: 10.9% +- 2.5%
CHG_Kotias: 14.8% +- 4.2%
Tarim_EMBA1: 21.3% +- 1.5%
pvalue: 0.18
Result File
I also try 2 other models, with EHG and Khvalynsk (75% EHG) as additional sources. Both of the results show Sarazm doesnt need EHG input, the coefficients are small (2%, with Std Error 3%).
EHG Result and Khvalynsk result
I think I have convincingly proved here that EHG ancestry is not required for Sarazm, and since Steppe_eneolithic and Steppe_EMBA both have significant EHG ancestry, theyre not sources for Sarazm.
3. Now let's get on with the Steppe_EMBA ancestry, which is younger than the Sarazm samples.
The model EHG+CHG+Tarim_EMBA1 fails with p-value 1.1e-8. Result here. It fails because of following generated DStats, which tells us about why our models fail when compared to the actual target.
gendstat: CHG_Satsurblia PPN -4.014
gendstat: CHG_Satsurblia Iran_GanjDareh_N -5.851
gendstat: CHG_Satsurblia Turkey_N -4.677
There's too much CHG and too little IranN, PPN and Turkey_N. There are other gendstats too which tell us the same thing.
So, in next model I add IranN as source. It fails with p-value 0.004.
Result
The worst gendstat tells us Turkey_N is needed
gendstat: Mbuti.DG Turkey_N -2.812
So I add Turkey_N as well. This model passes
left pops:
Steppe_EMBA
EHG: 32 +- 2.4%
CHG_Kotias: 14.4 +- 3.2%
Tarim_EMBA1: 43.5 +- 2.5%
Iran_GanjDareh_N: 5.7 +- 5.4%
Turkey_N: 4.3 +- 2.9%
pvalue: 0.12
Result
Where could this minor IranN + Turkey_N come from? Either from Sarazm like population directly or via Steppe Eneolithic, those are only 2 choices.
So, I model STeppe_EMBA with Sarazm as extra source. It passes.
left pops:
Steppe_EMBA
EHG: 32.7 +- 1.8%
CHG_Kotias: 7.3 +- 2.3%
Tarim_EMBA1: 36.6 +- 2%
Tajikistan_C_Sarazm: 23.4 +- 3.5%
pvalue: 0.20
Result file
So the relationship is actually exactly the opposite, Sarazm doesnt need EHG source, but Steppe_EMBA needs and IranN + Turkey_N source. In fact, Sarazm acts as a source for both Steppe_EN and Steppe_EMBA. And also for later pops like Yamnaya, Sintashta - indirectly.
Note: For Steppe_EMBA I have clubbed 1 high SNP sample from Kumsay (as all 4 are related), and 2 from Mereke.
Also, its just not me who has figured out that Sarazm source is required for Steppe_EMBA. Davidski has already modeled Steppe_Maykop with Sarazm.
Steppe_Maykop has same ancestry profile as Kumsay or Mereke, and the cluster together on PCA.
Eurogenes- The Steppe Maykop Enigma
By next weekend we should have some answers about PIE at least. Reich claims there was two West Asian admixtures in Yamnaya. Wonder what those west asian admixtures were but have you noticed any CHG or West Asian admixture in EHG? One of the EHG carries J1 haplogroup, this could be some kind of first wave west asian admixture event.
The 2 admixture are already known.
1st one is seen in khvalynsk, chg like admixture upto 20%.
2nd one adds more iran/chg admixture, as seen in Progress samples.
Are you suggesting second one which adds Iran/CHG is more Hotu-type ancestry + CHG?
Some unconfirmed leaks from the study about Iron Age samples from Iran "16 samples from Hasanlu in the northwest of Iran, all of them R1b with no EHG ancestry. The samples are all R1b Z2103"
2nd wave has sarazm like ancestry imo.
No EHG in Hasanlu is interesting. The IA samples are R1b too. What dates?
it makes sense only if 16 samples hasanlu R1b are from chalcolithic.( but some confusion that they are from 971BCE IA. )
Without old R1a clades, like R1b, found in North-west Iran, no wonder Reich lab sticking with Indo-aryan 'incursions' were later into indian subcontinent via Steppe ancestry & particularly R1a-z93.
It would be interesting these chalcolithic samples of Hasanlu had IndiaN!
Well Hasanlu_IA single sample is hard to model (coz of shotgun sequencing perhaps), but the nearby Hajji_firuz_IA 1100bce sample is very similar to hasanlu_IA.
Can be modeled as Hajji_C + Hajji_BA + 25% BMAC like. Hajji_B is 2400bce, and has part yamnaya ancestry.
Result
Chariots on a cup have been found at hasanlu, 2400bce is too old for yamnaya to have sent that to w iran. So BMAC/E iran is a good bet for the source.
"It would be interesting these chalcolithic samples of Hasanlu had IndiaN!"
@Muthu, given ashish/vasistha and maiyer etl al preprint modeling revision of IVCp where IVCp samples were found to have ANF(Anatolian Farmer ancestry) and their qpGraph allowing 4 admixture events putting the question mark on the early holocene split of IndianN(or IVC iranaian like ancestry) and Ganj Dareh_N, should we still call it as India_N ? Isn't it supposed antiquity in indian subcontinent is under question mark ?
If IVC's major source of west eurasian ancestry was via some south central Asian population(or some population in vicinity of it), that would again mean the IA(or II) languages was still intrusive to indian subcontinent. Only difference being that they arrived possibly around early chalcolithic period.
@Piyush,
yes, "given ashish/vasistha and maiyer etl al preprint modeling revision of IVCp", it is now more likely that this "IndiaN" ancestry arrived around early chalcolithic period before Mature harappan period.
but given the very small size(but not trivial) of the ANF ancestry contribution to IVCp, we can not rule the possibility of the interactions from the Anatolia AFTER "IndiaN" spread from NW India/Afghanistan.
normally we only think about simple one way flow of ancestry, but here we talk about few thousand years of time period. And that too possibly before Iran HG related and AASI fully mixed. there could be multiple admixtures both ways.
"but given the very small size(but not trivial) of the ANF ancestry contribution to IVCp, we can not rule the possibility of the interactions from the Anatolia AFTER "IndiaN" spread from NW India/Afghanistan."
@Muthu, 7%-8% ANF is in distal modeling, it does not mean that unadmixed ANF farmers directly crossed Iranian highlands and mixed with Indus neolithic people directly in say 4500 BCE. What it means is that the proximal source of the ANF in IVCp is from an already admixed source. That admixed source would mostly likely be from South-Central Asia or iranian plateau going by Ashish's proximal models here https://a-genetics.blogspot.com/2022/08/ivcandswat.html . IVCp cluster seems to get more than half of its ancestry from Tepe_Anau, Bustan_En like sources. Of course, the models may get revised with future aDNA but at this moment, seems likely that there might have been a significant plateau iranian or SC Asian admixture into IVC.
Lumping NW India with afghanistan is a bit problematic imo. It's more like a gateway between Central Asia and Indian subcontinent.
Yes Piyush, a good chunk of IVC ancestry coming from SC Asia remains likely, given the anatolian component in IVC.
However, this still does not mean that ALL OF IranN related ancestry in IVC came from SC Asia. Maier et al 2022 in their various 4 admixture graphs presents cases in which IranN related ancestry comes in via both IndiaN (which splits before Ganj Dareh) as well as a Tepe Hissar related source.
See graphs here
A drawback of Maier's graphs, like Narasimhan's graphs in SHinde et al is that they both dont use Tarim_EMBA as a population in the graph, and we know that admixture is present in IVC.
On the other hand, the drawback of my previous graphs about IndiaN (in the steppe eneolithic context) is that I don't include AnatoliaN population to reduce complexity.
5000bce samples from India are required to sort this issue.
exactly, we'd be able to dismiss any theory convincingly only after we have good quality samples from Neolithic, Chalcolithic and Bronze Age samples from the proper IVC
"By sequencing 727 ancient individuals from the Southern Arc (Anatolia and neighbors in Southeastern Europe and West Asia) over 10,000 years, we contextualize its Chalcolithic and Bronze Ages (~5000-1000 BCE), when extensive gene flow entangled it with the Eurasian steppe. At least two streams of migration transmitted Caucasus and Anatolian/Levantine ancestry northward, contributing to Yamnaya steppe pastoralists who then spread southwards: into the Balkans, and across the Caucasus into Armenia, where they left numerous patrilineal descendants. Anatolia was transformed by intra-West Asian gene flow, with negligible impact of the later Yamnaya migrations. This contrasts with all other regions where Indo-European languages were spoken, suggesting that the homeland of the Indo-Anatolian language family was in West Asia, with only secondary dispersals of non-Anatolian Indo-Europeans from the steppe."
https://www.universiteitleiden.nl/en/events/2022/09/the-genetic-history-of-the-southern-arc-a-bridge-between-west-asia-and-europe
Where do the Greeks como from? Anatolia or Steppe(going around the black sea)?
Idk yet. Earlier had done some rough analysis and Conclusion was that North greece saw some steppe input, and south greece saw west asian input. Due to graeco armenian linguistic branhch, i think the input from west asia at least influenced greek to some extent.
Update:
Ran 495 rotating qpAdm models on IVCp, 12 candidates, 4 sources at a time. I made a new python script to collate all the results in one file.
Only 1 model out of 495, which we have discussed above, passes.
Google sheet link to the model output here, collated in one place. Easily filterable.
Post a Comment