In this post, I analyze the date of steppe admixture with local ancestry in Swat Valley Iron age samples (avg 910 BCE) and RoopkundA (avg 850 CE) samples from Uttarakhand, India. These are the only 2 sets of good quality ancient DNA samples from the Indian subcontinent, and therefore extremely important to study the genetic changes in the subcontinent.
In one of my earlier posts, I analyzed all 23 RoopkundA samples and show that in that set of 13 male samples, the steppe component is not correlated with Y-HG R1a1a, instead, it is correlated with Y-hg J2.
Before I proceed with the admixture dating, there is something important I need to address regarding Swat Valley IA (SPGT) samples. With the Tarim basin aDna from 1900 BCE being published, I now see the need to change the model for SPGT. The informal G25 models show affinity to Tarim Basin ancestry and reduced Sintashta admixture. I caution though that this is based on informal modeling, and I am yet to do formal qpAdm models for SPGT with this new idea.
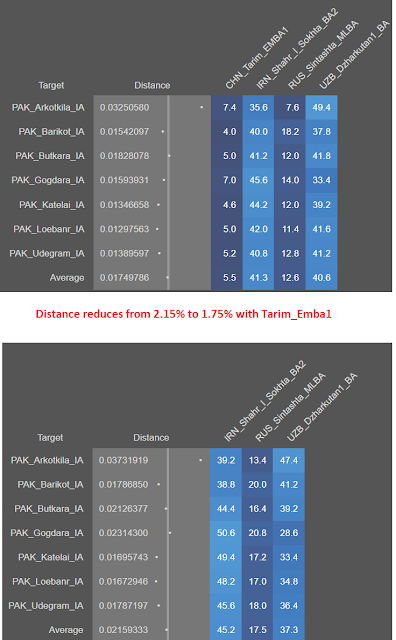
I model Swat samples in G25 using the Vahaduo tool with the following sources
1. Tarim_EMBA1 - Xiaohe culture China 1900 BCE
2. Shahr_Sokhta_BA2 - Indus periphery samples from Iran (3000-2500 BCE)
3. Steppe - Sintashta MLBA 2000 BCE
4. Dzharkutan_BA - BMAC samples (1900-1500 BCE)
The site from Swat valley called Aligrama is different from the others. It shows almost no steppe ancestry even by 850 BCE. I call these 3 Aligrama samples Pak_lowsteppe later in this post, along with select other samples.
 |
| Aligrama_IA shows elevated Tarim_EMBA and almost nil Steppe ancestry |
 | |
|
All the other Swat_IA/SPGT labels from 800-1100 BCE show around 12% steppe ancestry in this new model. This is down from 18-20% earlier, Tarim_EMBA seems to have eaten 6% from steppe ancestry.
Steppe ancestry in Swat medieval samples (1100 CE and 1700CE) jumps to 30+%, like moderns from the region today. I will come back to this later.
As shown in my previous post on Roopkund, BMAC ancestry is not needed to model Roopkund individuals, thereby eliminating Swat ancestry as a source for 800 CE Indians, Swat ancestry being rich in BMAC ancestry.
Admixture Dating using DATES for Swat_IA
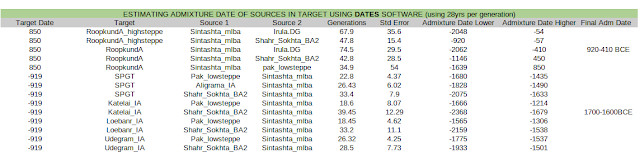
DATES is a tool developed to estimate the date of admixture of 2 specified sources in a target population. The theory is detailed in Narasimhan et al 2019 supplement section 6.2.1.
Using this tool, Narasimhan et al give the date of steppe admixture in Swat_IA as 1815-1479 BCE (S6.2.4.2 Narasimhan et al supplement)
I improve the methodology used in that paper by including multiple DATES models using multiple sources for local ancestry (Shahr_Sokhta_BA2, Aligrama_IA, Pak_lowsteppe) and using the common range of the 3 outputs to narrow the final range of admixture dates. Pak_lowsteppe is a subset of 8 samples from Swat_IA, including the 3 from Aligrama, with minimal to no steppe). The parameters for DATES were kept at default, the param file can be seen in the supplementary data linked in the relevant section below.
I confirm Narasimhan's dates but narrow it down to 1700-1600 BCE for the 12% avg. steppe admixture into Swat.
Admixture Dating using DATES for RoopkundA
As detailed in my Roopkund post, it was found that 23 of these Roopkund lake Individuals were Indians seemingly from differing jAtis and geographies within India. 10 were women, and 13 were men. They all died between 700CE and 900CE, possibly in separate incidents. It is likely that these people were going on a 'teerth' to Nanda Devi temple and had an accident/bad weather during the mountainous trek. So far, it is the only high coverage group of ancient DNA that we have from India.
What makes this group A so fascinating is that all these individuals are unrelated and are like a snapshot of the whole of India at that particular time (800CE).
I ran DATES for RoopkundA with Sintashta_MLBA and ShahrSokhta_BA2/Irula as sources to find out when these two mixed.
I also ran the algorithm on only the samples in Roopkund with >20% steppe ancestry to eliminate low steppe samples as a possible source of error. The results show that the admixture of steppe with the IVC-like component occurred 53 generations before the samples dates, with std errors and 28 yrs per generation (Moorjani et al 2016), gives admixture date between 920-410 BCE.
 |
| DATES shows Steppe admixture range for Swat & Roopkund, 68% confidence interval |
Conclusions
1. With the use of the latest Tarim bronze age genomes as a source in the model, the Steppe component in iron age Swat reduces to around 12%.
2. The steppe component entered ancestors of Swat samples between 1700-1600 BCE. This date agrees with the date of the Narasimhan et al paper, only the range is more narrow. It is not clear if and when this same spurt of ancestry entered the Indian mainland.
3. The steppe ancestry admixed with ancestors of RoopkundA samples between 920-410 BCE. Many of these samples look like brahmins on the PCA. In that case, does this late date justify the theory that the steppe component brought languages to brahmins and thus to India? It does not.
4. A big chunk of steppe ancestry in modern Indians and Pakistanis must have arrived post the Iron Age, as evidenced by DATES and Pakistan_medieval ancestry.
Edited to add:
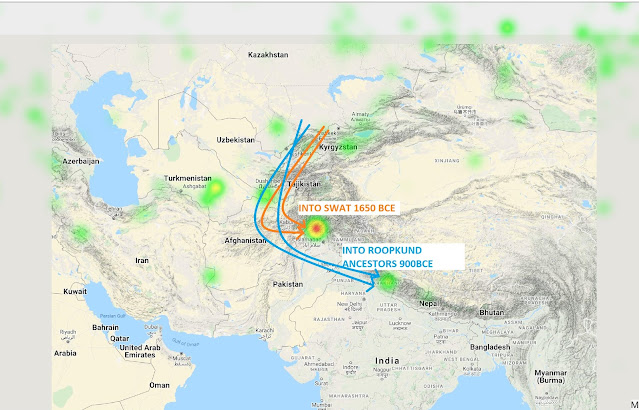
5. There seem to be 2 pulses of steppe ancestry into the Indian subcontinent. First came admixed with BMAC ancestry around 1650BCE, localized to the region around Swat valley.
The second pulse came deeper into India post 900BCE, this time with minimal to no BMAC ancestry.
In my view, it is possible that this steppe admixed population was residing in current Pakistan, Afghanistan, and NW fringes from 900-400BCE, and this ancestry made way into Indian populations through the annexation of these regions by the Mauryans (320-180 BCE) who enabled mass movements within its large kingdom. The population of India during Mauryan times is estimated to be 15-30 Million.
References
Zhang, F., Ning, C., Scott, A. et al. The genomic origins of the Bronze Age Tarim Basin mummies. Nature 599, 256–261 (2021). https://doi.org/10.1038/s41586-021-04052-7
Harney, É., Nayak, A., Patterson, N. et al. Ancient DNA from the skeletons of Roopkund Lake reveals Mediterranean migrants in India. Nat Commun 10, 3670 (2019). https://doi.org/10.1038/s41467-019-11357-9
Narasimhan VM, Patterson N, Moorjani P, et al. The formation of human populations in South and Central Asia. Science. 2019;365(6457):eaat7487. doi:10.1126/science.aat7487
Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, Reich D. A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc Natl Acad Sci U S A. 2016;113(20):5652-5657. doi:10.1073/pnas.1514696113
Supplementary Data
Results of various DATES models can be found here. In each subfolder, the output file with the No of generations and std errors has the extension .jout. The model can be identified by the name of the subfolder. On Linux, all these files can be viewed by a text editor, on windows these files can be downloaded and the extension changed to .txt so it can be viewed in notepad.
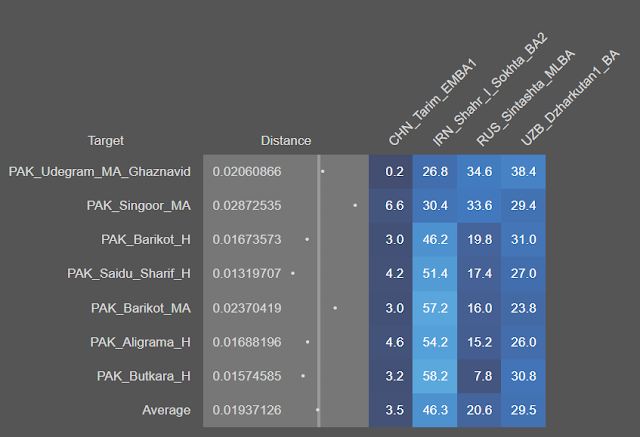
And if that doesn't point to enrichment through Parathians Scythians and Kushanas then what Will?
ReplyDeleteWhen will other Indians grow some grey matter and start understanding things......
Post 1000bce as far as Magadh, Prakrits or their proto versions were being spoken. If steppe ancestry came post 1000bce and steppe=IE then this will give ammunition to neo Buddhists who claim Sanskrit is younger to Prakrit and Buddhism is the religion of India lol. Which will also raise another likely question as to whether Harappa was Prakrit speaking.
ReplyDeleteBoth unlikely hence Harappa was indo-European speaking from 3500bce onwards with likely hood of other languages also coexisting which died a natural death. People from steppe came and assimilated into Indian society after all India was the mother of all steppe cultures.
Steppe IE-ising India post 1000bce is chronologically, archaeologically, linguistically impossible.
ReplyDeleteExactly!
Delete"What is the likely hood that apabrahmsha originated out of these foreign tribes trying to assimilate into the Indian culture?"
ReplyDeleteYou ask me such questions that I don't even know how to begin to answer.
hehe
DeleteI have way too many question and way too many models for way too many theories but nobody to ask and get counter views.
So I go around asking online trying to get answers to whether it's right or wrong or somewhere in between.
"The Indians referred to all alien cultures and races that were less civilized in ancient times as 'Mleccha' or barbarians. Among the tribes termed Mleccha were Sakas, Hunas, Yavanas, Kambojas, Pahlavas, Bahlikas and Rishikas. The Amarakosha described the Kiratas, Khasas and Pulindas as the Mleccha Jātis. Indo-Greeks, Scythians, Kushanas and Arabs were also mlecchas."
ReplyDeleteAin't it interesting that Sanskrit speaking Aryas never considered Dravidians as mleccha. They probably considered them their own family unlike some modern Dravidian groups who hate us northies like hell.
While Kurganists go gaga over horse, wheel and Carts ...
ReplyDeleteWe should never forget that our ancestors were conducting trade with Mesopotamia on ships and through 3000 km land route much before steppe people managed to tame horse.
Can you do it for Britannia? It is said that there was a replacement of population circa 2600BC, but there is no meaningful cultural change, except in the very very south, until the middle of 2nd millennium BCE. This is when the Halstatt culture enters the island, and this is the the first culture that is really continuous with the later Celtic.
ReplyDeleteThis is a situation which is the contrary of India, where you see continuity from at least the early 3rd millennium, with the the "invasion" happening in the middle of the 2nd.
"Steppe IE-ising India post 1000bce is chronologically, archaeologically, linguistically impossible."
ReplyDeleteBut Narasimhan, in the Questions & Answers of his 2021 lecture found in youtube, go by that way. He seems to suggest that Steppe ancestry entered India around Mahabharata's times (c. 900 BCE):
https://www.youtube.com/watch?v=K5hGa6ahSVc
(see from 1:12:00 approximately)
Maybe, paradojically, Narasimhan reached a similar conclusion than you vAsiSTha.
Post 1000bce steppe component accompany Iranian languages. Indians don't speak Iranian languages hence Indo-Europeanization post 1000bce is impossible.
DeleteWow, so what was india speaking pre Mahabharata if steppe and IE only arrived during Mahabharata? oldest mandala of RV is at least 500-800 yrs older than Mbh.
ReplyDelete@Vasistha
ReplyDeleteMathematically interesting to say the least.
The DATES tool - used by Narasimhan - is it possible to get some regression analysis of its behaviour? I am making a statement below -
The closer an actual aDNA find is to the direction of date of admixture, the closer will be the predicted date of admixture. So I am thinking that DATES is not linear in its prediction but is rigged to display a polynomial regression with respect to the independent variables.
Or is it totally wrong?
DATES indeed presents linear regression to the offered data and it is merely the new data (Roopkund) and the treatment (Swat vs Roopkund) that presents a different date?
@bruin
ReplyDeleteThe number of generations to admixture is a number that is solved by getting a solution to an exponential decay function.
Under admixture of 2 sources into target model, the covariance of a specific residual function ((function of allele frequencies at a particular snp in all 3 samples - target, s1, s2) ) at different SNPs i & j, where i & j are separated by distance d in centimorgans, decays at an exponential rate as a function of d. The older the admixture, the faster the decay.
So, from the samples inputted, the actual graph of decay is plotted and fitted into an exponential decay function, and from that function Generations and std error of generations are computed and provided as output.
The method has been calibrated using radio carbon-dated paleolithic samples.
ReplyDeleteSo no, a modern sample set will not give 1000yr later admixture dates than roopkund just because it is modern.
ReplyDeleteThanks, Vasistha
ReplyDelete"....the older the admixture, the faster the decay"
From the above statement, I do see it as a specific case of application of the "half-life" concept of radioactive substances.
However one major catch - the radioactive substance has no "memory" and no "cognizance" of surrounding environment. It will decay at the same rate whether it is a single atom or 1 million kgs.
Whereas the autosomal component will decay or not decay depending on the availability of mating partners with similar autosomal components.
Narasimhan's tool will have some floor built in to account for these assumptions. I am looking for that specifically. If the Steppes intrusion into India happened -
Like a steady trickle over several centuries
OR
One "big-bang" event
The tool should behave differently in both cases, I am sure of it. You wrote that you ignored datapoints with low Steppes admixture to eliminate error. I think that error is a manifestation of the tool's limitations.
All these 500bce-500ce invasions/migrations from steppes/Central Asia were mostly limited to Afghanistan Pakistan and North West India.
DeleteNone of these incursions made there way south of Vindhyas.
@daniel
ReplyDeleteI'll see when I can do DATES analysis for european populations.
For Britain, I see at least 2 large-scale population turnovers (i mean almost complete replacement).
1. One post 4500bce when anatolian farmers invaded and almost replaced WHG ancestry.
2. Post CW and BB cultures around 2000bce when this anatolian ancestry was replaced by BB
@Bruin
ReplyDelete"You wrote that you ignored data points with low Steppes admixture to eliminate error. I think that error is a manifestation of the tool's limitations."
I did not ignore them. I computed admixture dates with high steppe RoopkundA as well as all RoopkundA. Results were similar and there was no error. Results can be seen in the last pic in the post.
"Like a steady trickle over several centuries
OR
One "big-bang" event"
This tool assumes a one-shot admixture, however in reality it gives the average of steady trickle duration, the midpoint so to say.
"Whereas the autosomal component will decay or not decay depending on the availability of mating partners with similar autosomal components."
We already know that there was the availability of mating partners with steppe component, otherwise, the target samples would have ~nil steppe and I wouldn't have done this analysis.
@vasistha
ReplyDelete"...This tool assumes a one-shot admixture, however in reality it gives the average of steady trickle duration, the midpoint so to say."
A seven generation decay time (175-195 years) is the standard for disappearance of autosomal components in the absence of similar mating partners. This equals to almost 9 to 10 cycles between the Roopkund_A people and their supposed Steppes ancestor intrusion (900 BCE per your calculations).
Its a lot. Frankly, for me, that stretches very thin the viability of this calculation. But I understand this is the best we can do with the current available Indian aDNA.
But I definitely think that this has much higher valence than Vagheesh's use of modern Indians for Steppes admixture. So his own tool drags the admixture event forward in time when older datasets are used. Thats the biggest takeaway for me.Thanks again for the article
"A seven generation decay time (175-195 years) is the standard for disappearance of autosomal components in the absence of similar mating partners. This equals to almost 9 to 10 cycles between the Roopkund_A people and their supposed Steppes ancestor intrusion (900 BCE per your calculations).
ReplyDeleteIts a lot. Frankly, for me, that stretches very thin the viability of this calculation"
How does it do so when most roopkundA samples have steppe ancestry?
How is loss of autosomal ancestry a concern here when all have steppe ancestry (except 2 from roopkund)?
What this data suggests is that admixture between steppe and local ancestry happened between 900 and 400 BCE with the avg point at 650 BCE.
My analysis is confirming vagheesh's analysis for swat IA as target. The 12% avg steppe did reach there around 1650BCE. I only narrow the range further. So him using moderns as source and me using ancients didn't really change the results much.
ReplyDeleteEarly admixture would have happened in the iamc itself from harappan outposts like shortugai. Can we do similar analysis for iamc?
ReplyDeletePopulations? Also I would think the earlier iamc pops would be more xiaohe like with later entry of sintashta.. we should check that admix date too.
Now coming to models. Shortugai shows a northward trade and cultural influence. It’s unlikely that people with the sintashta component in iamc had continuity with languages spoken by andronovo people.
Over time it would make sense for these groups to adopt either Indian or Iranian dialects for trade And upward mobility.
During peaceful times there would be female exogamy including that of elite females both ways. Demographically bmac jeitun and harappan populations were very large and it would be those languages which would dominate.
Kushaan era power grab could not have happened in vacuum and must have been preceded by destabilization and some demographic events. Let’s also remember that Kushaan were preceded by Alexander’s armies, which would have fielded a large number of bactrian and central Asian mercenaries . They would be seeking to usurp power and alter the equilibrium.
@vasistha
ReplyDeleteI am going to read both the Vagheesh paper and yours again. My initial reading was that you have sharpened the admixture date from what Narasimhan postulated. Apparently it is not.
At least Narasimhan implied that the Steppes component entered India by 2000-1500 BCE, but the fine print apparently is only Swat Valley.
@bruin
ReplyDeleteIndeed,My analysis shows that there are 2 spurts of steppe ancestry. First in swat valley around 1650bce (agreeing with Narasimhan, only narrower range than his 1810-1480bce) and second in mainland India around 650 bce average.
I don't remember if Narasimhan mentioned the fine print about his dates being only for swat, but if he didn't do so then he is getting ahead of the data.
I have conclusively shown using his own method that the steppe admixture date for roopkund samples is much younger than swat and that the range is non overlapping.
Is it the same "Ghost population" which Narasimhan et al mentioned in his paper?
ReplyDelete
ReplyDeleteEdited to add:
5. There seem to be 2 pulses of steppe ancestry into the Indian subcontinent. First came admixed with BMAC ancestry around 1650BCE, localized to the region around Swat valley.
The second pulse came deeper into India post 900BCE, this time with minimal to no BMAC ancestry.
In my view, it is possible that this steppe admixed population was residing in current Pakistan, Afghanistan, and NW fringes from 900-400BCE, and this ancestry made way into Indian populations through the annexation of these regions by the Mauryans (320-180 BCE) who enabled mass movements within its large kingdom. The population of India during Mauryan times is estimated to be 15-30 Million
Indian Buddhist monks started moving to central Asia and China during the mauryan era.
ReplyDelete@vasistha
ReplyDeleteIf they were added by Mauryans so late, they might have had a somewhat different cultural identity and might even be identified by Greeks. Who were they?
If I am not wrong as per Greek accounts some Indians were white while others were dark not like the Ethiopians, tall and sturdy.
DeleteLet me find that pdf I read sometime back.
Sorry still not able to find that file but here is another interesting info:
Delete"Arrian, in the commencement of his “Indica,” mentions the Assaceni and the Astaceni as Indian nations in the mountains between the Indus and the Cophenes; but he distinguishes them from the other Indians as being less in size and fairer in complexion."
I make the claim because
ReplyDelete1. steppe ancestry can only mix with IVC ancestry first in the northwest.
2. the mixing happened avg 650 BCE.
3. the only pan India kingdom capable of enabling rapid transmission from Afghanistan to Bengal to Karnataka is the Mauryan kingdom. It may have started prior to that too who knows. maybe achaemenid conquest on NW in 550bce sent them into mainland. we will see.
Yup!
DeleteBTW the myth about Greeks in india before Alex's invasion, is it proven myth or is there any truth to it?
Can't say @ashishkaul
ReplyDelete@kaul
ReplyDeleteAssaceni are swat people only, only different timeperiod
Yup! Too long a timegap.
DeleteMany people had moved into the region by the Alexander's time.
This comment has been removed by the author.
Delete@Daniel de França MTd2
ReplyDeleteFinally found
Greek image of Indian society
https://ejournals.epublishing.ekt.gr/index.php/makedonika/article/viewFile/6094/5832.pdf
A Steppe entry of 800-500 BCE plays perfectly into the non-mainstream long chronology of indian history. Thats when the first Scythians entered India.
ReplyDeleteI would like to know more about this "non-mainstream long chronology"
DeleteLinks please 🙏
yeah some of those high steppe roopkund samples do show some affinity to Wusun and kushana/kangju groups in G25. will analyze that later.
ReplyDelete> yeah some of those high steppe roopkund samples do show some affinity to Wusun and kushana/kangju groups in G25. will analyze that later.
ReplyDeleteExciting, I hope my hunch turns out right. If further analysis shows this is the case, this will be big piece of evidence in favor of Long Chronology.
@AshishKaull
ReplyDeleteSearch up Vedveer Arya. He has made his three books public on his academia.edu page. A lot of reading though.
@3rdacc He puts Alexander back to c. 950BCE. That's hard to believe because it is tied to Egypt's chronology, which is calibrated by C14 dating to the mainstream age. So, I can't take him seriously.
ReplyDeleteTypical dates for trade along the Silk Road start at 200 bc but evidence for dairying with xiaohe mummies show that there was trade and transmission of ideas along the IAMC route even in 2000 bc.
ReplyDeleteThere was transmission of barley and sheet along IAMC since 3000bce. You see grains from SC asia in begash/tasbas at 2600bce along with first recorded cremation in the region.
ReplyDeleteHi vasistha,
ReplyDeleteSome questions
1) Given how genetically isolated tarim_emba samples were, it's doubtful that a population like that would have contributed directly to the swat valley iron age pop ! Do swat valley ancients scoring some tarim_EMBA like ancestry mean that these ancients need some extra ANE/WSHG like ancestry than what's accounted for in the Indus Peripheries ? The 'need for extra ANE/WSHG on top IVCp' theme is appearing again and again
2) Regarding dates model, I remember I commented about admixture dates of various high steppe castes provided in Narasimhan's paper in an earlier blog of yours but you seemed to brush that off due to the large range. Here, you have assumed 2 source mixture model (Sintastha_MLBA with either Irula or Shahr_Sokhta_BA2). Is such a two source model valid from a historical point of view ? Shouldn't it be a three source model or either Sintastha+IVCp like pop mixing Irula like pop ?
3) I was browsing through anthrogenica and I saw pegasus's comment regarding your post. He commented that - "Gujarati Brahmins, Rors get a date between 1200-1000 BCE, which perfectly coincides with PGW. ". It seems that they have run DATES on Ror samples and get earlier dates for steppe admixture. What are your thoughts on this ?
> He commented that - "Gujarati Brahmins, Rors get a date between 1200-1000 BCE, which perfectly coincides with PGW.
ReplyDeleteNo, newer carbon dating of PGW brings it before 2000 BCE.
https://www.academia.edu/36067852/Painted_Grey_Ware_Culture_Changing_Perspectives
"the earliest phase might be considered to have begun about 2300 BCE and the late phase as late as 700 BCE. If this is actually the scenario, then the PGW might be the longest surviving pottery tradition in Indian archaeological history."
Per Pathak et al (Am J Human Genet, 2018) the Ror are genetic outliers in Northwest India who (1) display more components related to EHG,Anatolia-N, CHG, Steppe-EMBA & MLBA than any other NW Indian pop (2) Are genetically closer to SGPT & BMAC than other NW Indians (3) Surprisingly, are the ONLY South Asian group genetically closer to Anatolia-N than to Iran-N. The Ror are also known to have the highest freq of lactase persistance allele -13910*T than most other Indian popns by far.
ReplyDeleteThis scenario is consistent with an initial injection of Steppe ancestry on the N/W periphery of the IV that remained relatively contained (little intermixing with Iran-N Heavy locals) and whose remnants still persist in the form of some modern Ror, Jat, Gujjar, & Kamboj descendants... separate & apart from the injection of Steppe ancestry responsible for most of the currently observed Steppe component in Indians.
So that would tie in with a 2-step insertion of Steppe ancestry.
Re: DATES
ReplyDelete"Under admixture of 2 sources into target model, the covariance of a specific residual function ((function of allele frequencies at a particular snp in all 3 samples - target, s1, s2) ) at different SNPs i & j, where i & j are separated by distance d in centimorgans, decays at an exponential rate as a function of d. The older the admixture, the faster the decay"
This appears to be premised on the concept of linkage disequilibrium. As more generations pass, the size of "Identical by Descent" blocks (in centiMorgans) becomes smaller and smaller as recombination events accrue. A pair of distinct but nearby SNP loci (i,j) would be inherited together less and less frequently (hence, covariance would decay) as number of generations from the coalescent increased.
What are the inputs? Do you specify the SNPs i & j? Or does DATES software go through the whole eigenstrat database and iteratively average over multiple pairwise i and j that it selects automatically?
This comment has been removed by the author.
ReplyDeleteThis comment has been removed by the author.
ReplyDelete@tim
ReplyDeleteTarim_emba like ancestry was present in indus periphery formation, and some also likely came in post bronze along with steppe. Exact origin is unknown, though it looks related to tarimEmba.
2. Dates will give admixture date of the 2 provided sources in the target. Narasimhan used steppe + Telugu for swat as target, whereas we know that bmac is another source in swat.
The output of DATEs also tell us if the model is valid, ie the out out graph is an exponential decline. If the model is invalid it will just be flat.
3. I haven't run dates on gujarati Brahmins or rors, so can't say.
RoopkundA has 23 random samples from the era and therefore a good set to analyze.
What are the inputs? Do you specify the SNPs i & j? Or does DATES software go through the whole eigenstrat database and iteratively average over multiple pairwise i and j that it selects automatically?
ReplyDeleteThe inputs are the genotype file and the target + 2 sources, plus parameter file.
The software goes through all SNPs i & j at a distance of d=0.45centimorgan + delta d.. delta d is varied to plot the covariance at each point d on x axis.
The resulting curve is an exponential decay curve, and the output is calculated from its rate of decay.
"whose remnants still persist in the form of some modern Ror, Jat, Gujjar, & Kamboj descendants... "
ReplyDelete@Indospehere,
Gujjars and Kambojs don't have much steppe_MLBA ancestry, their primary west eurasian ancestry is from Iran_N + WSHG like source, something like a very low AASI IVC periphery like source .
"Modern homos have inherited more DNA from their ancestors than native breeders", says professor Ano Komo from Kagawa University, 86% of their ancestry alone descends from Bootygig culture (850-320BC), Dark room culture (5000BC-60BC), Back Door cradle (800-100BC), and Manhole.
ReplyDelete"The results were astonishing" said in regards to such endogamy remaining even with cultural and migrational shifts. L5379 mutation virtually absent in remains, while unique Haplogroup R1d1ck was found in over 93% of Bootygig-Manhole complex mummies and reimains in 87% of modern "happy men".
The female lineage, however, seems slightly more mixed. "Paternally, it's a direct descend"
Experts explain it as no contact with local breeders and the 14% average local admixture being female-mediated (gaga el. al) while the 6% foreign male lineages coming from a low-grade influx of semiheteros as observed by madonna el al.
"Even the foreign lineages have native proto-homoland mutations at some part of the Y chromosome" The speculation, however, still seems unthinkable given historical oppression of semiheteros, and them being called "bifrutas" (bi-fruits in spanish) which was also the name of a drink.
Mixing and kidnapping of local women was more documented in infamous Dark Room towards the latest stages.
There's been talk of this foreign minimal amount of male haplogroups coming from slaves, or poor local breeders dominated and forced to convert while some others srressvthat it was just a folklore tale or urban legend.