Y Haplogroups - A Primer
Y Chromosome haplogroup is a defined set of mutations in the (non-recombinant) portion of the DNA from male specific Y-chromosome. Y-Chr is passed down from father to son along with all the mutations that have been accumulated till that point. All the male descendants of a man will share the mutations in the Y-Chr of that man, plus all the additional mutations that have been collected in the generations between that man and the descendant on his specific male ancestral lineage chain. This feature helps us in identifying the most recent common paternal ancestor of a group of men and is a great tool for population geneticists. Do note that the same set of additional mutations cannot be created in two different men at the same time, the probability of that happening randomly is 0.
mtDNA haplogroups are mutations found in the mitochondrial DNA. Unlike autosomal chromosomes and Y chromosomes, mtDNA is found outside the cell nucleus. mtDNA is passed down from mother to children only, and therefore is informative only of the maternal lineage of a person.
In this article, we will discuss Y haplogroups which can only inform us of the paternal ancestry of males.
 |
| Y Hg simple tree: Source |
The nomenclature for assigning haplogroup varies, and the most commonly used notations today are ISOGG and YFull. ISOGG notations go like this:
R>R1>R1a>R1a1>R1a1a>R1a1a1>.......
R>R1>R1b>R1b1>R1b1a>R1b1b1>.......
R>R2>R2a>....
Here, each subclade has an additional set of mutations than the previous ancestor. So, R1a and R1b have accumulated a different set of mutations from a common ancestor with Y-hg clade R1 which sets their lineages apart from then onwards. The common paternal ancestor shared by all individuals in the R1a, R1b and R2-carrying individuals is an ancient man who carried the R haplogroup.
A similar notation is applied to other all other haplogroups.
Here, it is worthwhile to get into the terminology that will be used in the subsequent portions of this article, explained at a very basic level.
- SNP - single nucleotide polymorphism. Pronounced 'snips'. Each SNP represents a difference in a single DNA building block, called a nucleotide. Each SNP is found in a particular position of the genome (defined by physical position) and is given a name and a reference allele (A, C, T, or G). Harvard ancient DNA database currently provides samples in 1240K SNP resolution (across all chromosomes, in reality not all SNPs can be retrieved from ancient samples). Of these 1.24 million, 32670 SNPs are on the Y Chromosome.
- Mutation - for a sample, a SNP for which the sampled allele is different than the reference allele.
- eg. These are some of the mutations which define the Haplogroup R as per ISOGG.
- In the first line, SNP Names M207, Page37, and UTY2 are given to the unique rsID rs2032658 found at position 15581983 (Human genome reference build #37). A mutation is detected if the allele 'G' is found at this SNP instead of the reference allele 'A'. This mutation, among many others, defines the Haplogroup R.
- Positive SNP: If a mutation is found at a SNP. eg. 'M207+' implies that 'G' was found at the above-mentioned SNP.
- Negative SNP: If a mutation is not found at a SNP. eg. 'M207-' implies that the haplogroup is likely not 'R'.
- Haplogroup - a major branch, such as Haplogroup E or Haplogroup I
- Clade - from the Greek word klados, clade means branch, and subclade means a further branch. eg. I2 is a branch of Haplogroup I
- Terminal Hg - the branch with the youngest estimated age (age closest to the present) for a sample in the currently available database. For eg., if a sample is found positive for R1a1a, and no further mutations in the available database are found positive, the terminal Hg for the sample is marked as R1a1a. This is important as it helps us ascertain male movements in ancient times more precisely.
- Upstream - all SNPs which are a link in the chain of ancestors till a given mutation.
- Downstream - all SNPs which are descendants of the ancestor with a given mutation.
- Asterisk * - An asterisk after a subclade means that there are further mutations after the terminal Hg but no other available sample shares those additional mutations, therefore they remain unclassified.
- Plus + - Means a positive SNP, indicating that terminal Hg is either at this SNP or downstream of it. eg Z93+ includes terminal Hgs Z93, Z94, Z2124, Y3, Y2, L657 and everything else under Z93.
- Minus - : This means a negative SNP call, indicating that SNP and all downstream SNPs are negative as well.
R1a and R1b Found in ancient DNA
In the aDna database, R* is found in 22000BCE Siberia. So we can assume that R was prominent in the ANE people (Ancient North East). The ultimate origin of Hg R is from the subclades of Hg P. Hg P is widely present among Malaysians and Filipinos. So, the ultimate origin of R is SE Asia (Hallast et al 2021). The route taken to reach Siberia might have been via India (P-M1254 in Andamanese). There are also two P-P337+ samples found in the 30000BCE site of Yana in Siberia. So whatever the case is, the Y haplogroups reached Siberia from Southeast Asia very early.
Clade R2
Formation date ~26000BCE. Subclade R2 appears in the oldest samples from Iran (Ganj Dareh 8000BCE). R2 is also found in SC Eneolithic samples (oldest from that region), but absent from other Central Asian and Siberian sites. So, we can hypothesize that some R men migrated from Siberia to SC Asia/Iran/India where it mutated into R2 in one of their descendants and spread across the region. A more likely scenario is that on the route from SE Asia to Siberia, some R men spread in India/Iran and their descendants mutated into R2.
Clade R1b
R1b splits into R-L754 (R1b1, seen in 12000BCE Villabruna) and whose descendants are visible in the neolithic steppe as well as modern Europeans. The other split is into R-PH155 (R1b2) only seen in the east (Southern Central Asia and Tarim). Basal R1b* (pre PH155 - it has some mutations of PH155, but not all) is seen in a BMAC sample from Uzbekistan. Furthermore, all the 2000BCE Tarim Basin samples are R1b-PH155 or pre-PH155.
With this, we can hypothesize that R1b was born in Siberia/Central Asia from R1 and spread to Western Europe where it mutated into R-L754 and mutated locally into R-PH155 in central Asia.
Clade R1a
The oldest sample is a 10700 BCE man from NW Russia, with R-M459 (R1a1). R1a1 is also seen in the EHG hunter-gatherers (Karelia) and the Eneolithic steppe (Khvalynsk). Local continuity is also seen till the bronze age, where in Fatyanovo we see R-Z645 (R1a1a1b) and the descendant R-Z93 (R1a1a1b2).
The brother clade of R-Z93 is R-Z283 (R1a1a1b1), which is first found in Estonia Corded ware 2600BCE and now is primarily present among central and east Europeans.
The absence of R1a in every sample from Iran and South Central Asia till 2000 BCE makes it very likely that R1a is indeed not local to this region. The earliest sampled R1a-Z94 in this region is associated with autosomal steppe ancestry.
R-Z94 Tree
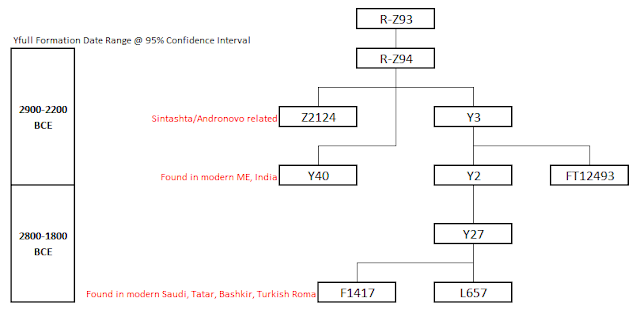 |
| Z94 tree as per Yfull |
Note: Formation dates are taken from Yfull. Here's their methodology for calculating age:
The second formula uses an assumed mutation rate of 144.41 years (0.8178*10-9, which is the average of the mutation rates of the ancient Anzick-1 sample and of a group of known genealogies, and an assumed age of 60 years for living providers of YFull samples.
Distribution of R1a and subclades in India by Region - Dr Chaubey Data
Of the 1196 samples analyzed from all over India, 23% belong to R1a or downstream. 16% belong to R-Y3+ (including L657) whereas 7% belong to the non-Y3 branch of R1a, and can be assumed to be of steppe origin, especially the ones which are Z2124+.
NW India has the highest % of R1a at 41%, of which the non-Y3 branches are 20% and the Y3+ samples are at 21%. Definite steppe impact is seen here with 13% Z2124+ samples. Ganga plains, central, east & south India have 23, 33, 20, and 17% R1a respectively. The bulk of that is Y3+, and only 4-10% is Y3-.
None of the samples from the central, east and Ganga plains show Z2124+. This difference in frequency in Z2124+ and Y3+ samples is important and I will come to it a bit later, just keep the numbers in mind.
Eurasian R1a distribution, from 11036 samples uploaded on Yfull as of Oct 2022
Let's have a look at some Specific Branches:
1. Z-93. This clade has many downstream subclades, 3 of which are minor and have a clear origin in the steppe region. These minor ones are
- R-BY226207 (most modern samples found in Tajikistan, Xinjiang, Italy & Kyrgyzstan. One ancient sample from Srubnaya_alakul bronze age steppe site as per Yfull).
- R-YP5585 (1 ancient sample from 1800BCE srubnaya_alakul, most modern samples from England, 1 from Kuwait and 1 from madhya Pradesh, India (community unknown)).
- R-FGC82884 (One notable ancient sample which falls downstream of this- DA382 Turkmenistan_IA 850BCE. Another ancient sample downstream is from bronze age Srubnaya context. Other modern samples are from Hungary, Poland, China, Kyrgyzstan, Kazakhstan, Russia, Arab countries).
2. Z94. Formed ~2600BCE from Z93. Z94 terminal has been found at steppe bronze age sites. It has also been found at Swat valley Pakistan iron age sites, and among some Saka samples, which makes sense given the steppe connection to SC Asia and Swat.
R-Z94 has 1 minor subclade and 2 main subclades. The minor subclade is R-Y40. The main subclades are Z2124 and Y3.
3. Z94>R-Y40. formation date ~2600BCE- No ancient sample under this subclade has yet been found. In terms of modern distribution, Yfull has downstream samples from a Tamil speaker, Singapore (likely a Tamil speaker), Bangladesh, Pakistan, Jordan, Saudi, Syria, Iraq, Italy (likey middle east connection), Xinjiang, Kuwait, Qatar, Kerala, Konkan, Gujarati, Tajikistan and Turkey. On FT DNA as well, samples with Y40+ are from Turkey, Saudi Arabia, Italy, Bulgaria (name Ismail), South India and Kuwait. In the Yfull data, this subclade of Z94 (and its downstream mutations) is only found in India, Pakistan, Bangladesh, China, the Middle East and Tajikistan even though its formation date is ~2600BCE. This mutation wasn't found in any of the 2200 Russian samples on Yfull. The birth of this subclade can be considered to be in the Indian subcontinent.
DISCUSSION
Notably, R1a-M780 occurs at high frequency in South Asia: India, Pakistan, Afghanistan, and the Himalayas. The group also occurs at >3% in some Iranian populations and is present at >30% in Roma from Croatia and Hungary, consistent with previous studies reporting the presence of R1a-Z93 in Roma.
There could also be bronze and Iron age connections from the Indian subcontinent to the middle east, however, we don't have ancient DNA from these regions to confirm or deny this.
 |
| Modern distribution of R-Y3+ (M780+). From Underhill et al 2015. Used under CC license |
The density (and diversity) of R-Y3+ is highest in the Ganga plains and East India, so that remains the likely region of expansion of this clade.
Did an army of macho Aryan men invade India and give the Vedic language and culture?
It is important to say at the outset that Y haplogroups do not biologically encode for the language spoken, that is a sociological outcome. In the modern world, there are R1a people who speak European languages, Indo-Aryan languages or Iranian languages but many R1a also speak Dravidian, Semitic, or Turkic languages. The most robust reason for the spread of language families like Indo-European is, as Bellwood 2020 puts it, "that they travelled originally in the mouths of migrating populations."
This means that autosomal ancestry change (agnostic to sex bias) is a stronger predictor of language change than Y-haplogroups. Unlike the Y chromosome, autosomal chromosomes (1-22) undergo recombination and both son and daughter receive ~50% admixture from each parent. So the level of external autosomal admixture coefficient can tell us about the ratio of migrants-to-locals, and analysis of the X & Y chromosomes of the samples can tell us about the sex ratio of those migrants.
The larger the autosomal ancestry impact from migrants, the more likely it is that there would be a large impact on the local languages of the region, and given a large enough migration (still a subjective %), there will be an eventual language replacement. This is exemplified by the presence of a high percentage of 'Yamnaya steppe' autosomal ancestry in Corded-ware bronze age cultures of Europe (>70% as per Scorrano et al 2021), which is the reason for the Indo-European languages in those parts of the world as per most researchers. At the same time, Basques are one of the few non-Indo-European speaking groups of modern Europe and they show lesser (~40%) Steppe ancestry but with 80+% Basque males being Y Hg R1b (Olalde et al 2019 and Luis et al 2021). R1b is considered to be a part of this steppe expansion west into Europe. So clearly in the case of Basques, high R1b frequency was not successful in replacing the language of all the Basque ancestors, but the autosomal ancestry gives us a better picture - lower (40%) in Basques vs 70%+ in IE-speaking Germany.
Without knowing the exact societal processes of a given part of the ancient world, the one statement which can withstand scrutiny is 'The larger the migration, the more are the chances of eventual replacement of the local language.' There are also recorded instances of languages being replaced due to migrant women, eg. the replacement and (almost) extinction of Badeshi by Torwali in Swat, Pakistan within a few generations due to Badeshi men marrying Torwali wives.
In South Asia, we detect eight lineage expansions dating to ~4.0–7.3 kya and involving haplogroups H1-M52, L-M11, and R1a-Z93. The most striking are expansions within R1a-Z93, ~4.0–4.5 kya. This time predates by a few centuries the collapse of the Indus Valley Civilization, associated by some with the historical migration of Indo-European speakers from the western steppes into the Indian sub-continent
Using previously reported calls on 1000 Genomes Project Y chromosomes, we observe that 62 out of the 221 South Asian males have an R1a Y chromosome corresponding to a ninety-five percent binomial confidence interval of 22-34% for Steppe MLBA ancestry on the entirely male line, which is significantly higher than the ninety-five percent confidence interval of 9-14% on the autosomes in the same set of individuals. These results shows the process of admixture of Central_Steppe_MLBA into the ancestors of the ANI was male biased, and reveal that the directionality of sex bias was opposite to the pattern observed for the contribution of Central_Steppe_MLBA to SPGT.
To put it simply, the authors are claiming that although the swat valley data says the opposite, modern data shows them that the steppe autosomal ancestry in modern South Asians must have come primarily via men from the steppe carrying R1a-Z93. The 221 South Asians are from four communities: Sri Lankan Tamil in the UK, Gujaratis in Houston, Telugu in the UK, and Punjabi in Lahore. Notice the erroneous assumption made here: All of Indian R1a is treated as Z93 without any of the nuances that they offer to other regions of the world. Eg. The recent 'Southern Arc' paper analyzes various subclades within R1b with nuance for Steppe, Armenia & NW Iran. The presence of M780 (aka Y3) and L657 in Indians was known since 2015, so that is not a valid reason to ignore the subclades of Z93.
The mechanism of Autosomal & Paternal ancestry transmission
Suppose that 1 R-Z94 man from the steppe migrated to Haryana and married a local woman there. They had 2 sons. The sons would have ~50% steppe ancestry and Z94 Y Hg. Suppose that both sons underwent separate mutation on their Y Hg and formed Z94>Y40 and Z94>Y3 respectively. Their male descendants would inherit these 2 subclades, but with each generation of marrying local women, eventually, the steppe ancestry would fall to ~0% in 100-120 years (50% >25% >12.5% >6.25% >3.125% >1.55% >~0)
1. The only 2 R1a males in the 850CE Roopkund_A cluster have zero to minimal steppe ancestry, whereas all the other male samples with much higher steppe ancestry are non R1a.2. Chenchu tribe of south India has ~25% R1a frequency but no steppe ancestry (Kivisild et al 2003)3. There's a CHG/Iran related Y hg J sample in Karelia, Russia which has only Eastern Hunter Gatherer (EHG) autosomal ancestry.
and many more.
However, the complete lack of association of R-haplogroup descendants and EHG ancestry in either Armenia or Iran is consistent with either a massive dilution of EHG ancestry in these populations resulting in the dissociation of Y chromosome lineages from autosomal ancestry over time, or with a scenario in which R-M269 was not associated with substantial EHG ancestry to begin with.

I was wondering why the Narasimhan paper doesn’t add up. The dominant r1a clade in India is L657, which is found earliest at 500 bce in China, not even in the Swat valley. So how can the swat valley steppe be source of Indian steppe ancestry
ReplyDeleteNot 500bce, L657 is found in 100bce Xinjiang. saomple id C3316 China_Xinjiang_Guanjingtai_IA
ReplyDeleteAs usual, a cutting edge analysis. I am reading the following sentence
ReplyDelete"Of the 1196 samples analyzed from all over India, 23% belong to R1a or downstream. 16% belong to R-Y3+ (including L657) whereas 7% belong to the non-Y3 branch of R1a, and can be assumed to be of steppe origin, especially the ones which are Z2124+. "
and trying to relate it to your Yfull box chart and unable to visualize how the R1-M780 relate to Y2/Y3/Y4. Another chart with only the India relavant Y at the top and then all the R's further down would help. Thank you.
R-M780 is another name for R-Y3.
ReplyDeleteAs per YFull, it is defined by mutations at 2 SNPs- Y3/F2597/M727 and Y26/M780.
If both these mutations are found, Yfull calls it R-Y3, whereas some research papers call it R-M780.
I looked at the YFupl for Y3, a lot of the people taken were Russian or middle eastern. How do you know of Y3 originated in India?
ReplyDelete"I looked at the YFull for Y3, a lot of the people taken were Russian or middle eastern. How do you know of Y3 originated in India?"
ReplyDeleteMiddle east has many samples, but it is the most oversampled region on Yfull. Check this map https://phylogeographer.com/yfull-world-sampling-rate-map/
only 30-40 ME samples out of 5000 are R-Y3+. This is less than 1%, and makes it unlikely that it was formed there. The Romani connection from India is a much better reason for R-Y3 presence in Arab world.
There are precisely 4 Russian samples, all with Z94>Y3>Y2>Y27>F1417, 2 of them are Tatars, 2 are Bashkirs. All 4 are Kipchak Turkic speakers and not Indo European. The earliest Kipchak inscriptions are about Buddhism. So a late spread due to Buddhist movements is the reason for this. These central asian Buddhists later became Turkic speakers. Alai Nura, Kyrgyzstan 400CE sample also falls downstream of this mutation. Samples with south asian admixture have been found in other sites near Alai Nura at the same time. This is the period when Buddhism was at its peak.
Good, I saw a video in a Youtube channel called World of Antiquity explaining the origin of Sanskrit using genetics. there was a link by someone to his channel explaining how genetics proves an archeic indo european presence in India. That was struck down due to the argument that ancient dna, somehow better, shows it is steppe peoplemoved after 2000 bce, not before. I wish they could see your work.
ReplyDeleteThat guy is wrong.
DeleteI always thought forcing the "male mediated steppe ancestry" on modern day South Asians was a deceiving layered lie. And for the Romanian explanation for presence of Y3 in Middle Easterners, do Romani themselves have this mutation (R-Y3+ and R-Y2-)?
ReplyDeleteOn Ftdna theres this Turkish Roma guy with Y3>Y2>Y27 L657-
ReplyDeleteE13038 Sarmaşık, Turkish Roma (Horahane), Babaeski Turkey R-M634
On Yfull, there's a Y3+ Y2- (Y3>FT12493) Turkish sample, more context is not given. He could possibly be Romani but cant say more.
There's an Omani as well as a Brazilian (Minas Gerais state) sample as well (Y3>FT12493>FT13945), with no extra known info.
The Brazilian sample could be Brazilian Romani, who have had presence in Minas Gerais after 1600ce, but also in other brazilian regions. https://en.wikipedia.org/wiki/Romani_people_in_Brazil
Older studies on Roma people don't differentiate between Y3, Y2 or L657, so theyre useless in this regard.
I am not sure R1A is the original IE.
ReplyDeleteJ1 and R1A appear together in a few places. Also IJ is kinda close. Why is IJ and R1a/b appearing so close to each other.
It looks like IJ left the IE homeland a long time ago, and then that homeland was left with R1A/B mostly, which is the last expansion we are seeing.
All of these HGs probably expanded out from the Gujarat region. R does seem intrusive into the IJ and G HGs which are phylogenetically closer and then R would have separated earlier than mixed in again or something.
I actually have J1-P58 but my ethnic group looking at 23andme is like 50% R1A and 50% J1
Shouldn't the dates of mutation be in general much older than they are attested? For example, for Z93 or Z94 to be found at a certain in certain individual, it is much more likely that it appeared much more in the past only to become common at a much later date.
ReplyDeleteIt's not like that a certain mutation will spread so soon, unless it yields some evolutionary advantage. So, Z93 could be a couple of thousand years old for a "random walk" spread. I don't have the mathematics means to calculate that, but I've never seen this point addressed.
The formation dates given are ranges at 95% confidence interval. eg. R-Z2124 date as per Yfull is 4900 - 4200 years ago at 95% confidence.
ReplyDeleteBut the Y hg has also been found in a sample dated to 4600years ago. So we can say that Z2124 was formed between 4900-4600 years ago.
Ding et al believe that the ages of R1a subclades are underestimated and that they should be older. If that is the case, then even Yfull formation dates should be pushed back, maybe by 10%.
@Vasistha but that is not what I asked. It seems to me that these dates refer to the error in estimating the oldest sample. What I want to know is that if, in this dating, they estimate the time it takes for a mutation to happen until it is common enough in the population so that it is not a "fluke".
ReplyDeleteFor all I know, it Z93 could appear, say, 4500BCE, in a very isolated population, but it and only centuries later diffuse. Or, maybe, it doesn't just diffuse by mere chance.
There is an error margin in the estimation, and if the clade formed much earlier than estimate then we should be able to find it somewhere in the ancient records.
ReplyDeleteOtherwise, the calculations are our best bet.
VAsishta You should send your work to the so called authorities and see if they will change their views. Also could you put this article on a YouTube video?
ReplyDelete"is present in 80% of Afghan samples. I think that this increase is a result of the brutal Ghaznavid invasion of Gandhara from Central Asia post 900CE."
ReplyDeleteHey Ashish, If I understand this correctly, are you claiming that increased frequency of R1a-Z2124 in Afghanistan is the result of Ghaznavid invasion ? If yes, then by extension, are you also implying that pashtuns, who are rich in R1-Z2124 and are the dominant ethnic group in afghanistan , received major patrilineal gene flow during this ghaznavid invasion ?
Folks were saying that the swat valley medieval ghaznavid era sample was very similar to modern day pashtuns.
"Hey Ashish, If I understand this correctly, are you claiming that increased frequency of R1a-Z2124 in Afghanistan is the result of Ghaznavid invasion ? If yes, then by extension, are you also implying that pashtuns, who are rich in R1-Z2124 and are the dominant ethnic group in afghanistan , received major patrilineal gene flow during this ghaznavid invasion ?
ReplyDeleteFolks were saying that the swat valley medieval ghaznavid era sample was very similar to modern day pashtuns."
There's a caveat, the 47 Afghan samples on Yfull are quite less in number. Maybe many of those individuals are related as well. So the 80% Z2124 on Yfull may actually not be accurate for Afg as a whole.
Yes, swat valley medieval samples do look similar to north_afg_pashtun on G25, but qpAdm is needed to validate that. What is certain is that there is increased steppe autosomal ancestry and R1a frequency after the Buddhist period, as seen in the medieval period samples in Pakistan.
R1a Frequency in Swat
IA - 2/44 R1a
Historical - 2/9 R1a
Medieval - 3/7 R1a
But maybe I'm not so off the mark. Underhill et al 2014 say
"R1a-Z2125 (Figure 3c) occurs at highest frequencies in Kyrgyzstan and in Afghan Pashtuns (>40%)."
Z2125 is downstream of Z2124, and is found at highest freq in Afghanistan as per Underhill, as is shown by Yfull data as well.
Update: Have added Yfull data for various European countries to the dashboard.
ReplyDeleteFor Kazakhstan, Uzbekistan, Tajikistan, Kyrgyzstan and Turkmenistan the Yfull samples are less in number. So I have also added 166 total samples from these countries from the following 2022 paper.
"Ancient Components and Recent Expansion in the Eurasian Heartland: Insights into the Revised Phylogeny of Y-Chromosomes from Central Asia" - Zhabagin et al. 2022 https://doi.org/10.3390/genes13101776
Hi Vasishtha
ReplyDeleteSo based on your modeling, you found that 1 or very few R1a1 people came from outside which resulted in 20% R1a1 presence in India.
What do you think of this paper? They are suggesting R1a1 possibly has founding behavior, I believe they are suggesting R1a1 roots are in India. Please comment.
https://www.nature.com/articles/jhg20082?fbclid=IwAR1ANswvT3Mv0qz2zNgJAMR3yF8cLwqBKE694jhtpZ2FMRC39lEZZBZdV-8
The entire ludicrous idea that R1a originates from Siberia or Northern Eurasia is completely without basis or merit, the claim that R1a originates from Siberia or Northern Eurasia COMES FROM ONE SINGLE INDIVIDUAL SAMPLE AND NOT MANY SAMPLES!! This is a case of cherry picking and pure obfuscation, and absence of evidence does not equal evidence of absence. IN REALITY IT SHOULD BE India-Southern Asia-Iran-------------------->>>>Central Asia------------->>>>Siberia and Northern Asia---------------------------->Then Europe. Haplogroups aren't the be all and end all for genetics and actually for individuals is a poor measurer of genetic estimation, however the genetic ancestral clade to Haplogroup R is Haplogroup P and P is an Indian South Asian origin genetic haplogroup and South East Asian so it's irrefutable that R had to have based on that reality alone originated from a South Asian-Indian and Iranian location origin. Secondly the OP is completely ignorant of the reality and fact not aside from Haplogroup "P" the ancestral genetic siblings of Haplogroup "R" such as Haplogroups Q, M, S, N are found oldest in either Indian South Asian origin population groups or had origin within South Asian populations in the distant past or found in geographic adjacent regions of India-Southern Asia region; for example Australasians-Aboriginals are the only people that today carry Y-Haplogroups "M" and "S" and not anyone else (definitely not Northern Eurasians), Haplogroups "O" and "N" which are the other genetic paternal lineages closest to "R" originated and migrated-split out of North-East India-South East Asia and it's accepted as fact that "O" and "N" originated from mainland South East Asia-Eastern Himilayas which is near historic South Asian "Onge" admixed populations or came about from Onge admixed population groups (accepting the premise that "Onge" represents ancestral SA HG populations which they certainly didn't) so it's pretty clear based on simple evidence that the R1 and "R" had to have Indian South Asian-Iranian origin source.
ReplyDeleteThirdly the entire premise of "R" originating from a Northern Eurasian location is based on cherry picking evidence and convenience sampling and absence of evidence does not equal evidence of absence. Because those locations had the privilege and finances and right conditions to uncover bodies and samples (single digit ones) researchers can cherry pick and claim X or Y genetic lineage originated from this or that location then ignore all contrary evidence which is what the OP is doing. Northern Eurasia and Central Asia today and in the past were barren cold wastelands incapable of hosting or housing large scale human populations in the past based on climatic conditions alone and still can't today specially to generate genetic diversity to originate genetic haplogroups. So, the idea that a major genetic lineage like "R" could originate from such desolate low population count regions such as Northern Eurasia especially right after the ice age is preposterous and unbelievable. So more non-sensical in-diot coolie non-sense and idiocy from the original OP anything other than a South to North migration is unbelievable.
Look at the Time to most recent common ancestor for R-M207 (Arunkumar et al), R-M198 (Mirabal et al), R-M17 (Swarkar Sharma et al, R-M780 (Underhill) and R-Z2125 (Underhill). The ratio between times for India versus Central Asia is 1.5 i.e Indian subclades are 1.5 times more ancient - i.e. they are pre-neolithic. R or R1 came to India during the LGM according to me. Wells et al and Mahal also said that it came in the pre-neolithic period. The Aryans just did not come in 1500 BCE, they did not even come during the neolithic they came to India as a result of the very bad climatic conditions during the LGM to India because it was the ony place with great climate even during LGM.
ReplyDelete