In 2019, a paper1 was published with the analysis of the remains of 38 individuals whose bones were found in Roopkund lake, Uttarakhand at an altitude of 5000m above sea level.
 |
| Context of Roopkund Lake. a Map showing the location of Roopkund Lake. The approximate route of the Nanda Devi Raj Jat pilgrimage relative to Roopkund Lake is shown in the inset. b Image of disarticulated skeletal elements scattered around the Roopkund Lake site. Photo by Himadri Sinha Roy. c Image of Roopkund Lake and surrounding mountains. Photo by Atish Waghwase |
13 of these skeletons were confirmed to have died around 1800CE in a single event. These 13 individuals had east Mediterranean ancestry, ie. they were foreigners. They were also accompanied by 1 person of Malaysian/Vietnamese descent.
On the contrary, it was found that 23 of these Individuals were Indians seemingly from differing jAtis and geographies within India. 10 were women, and 13 were men. They all died between 700CE and 900CE, possibly in separate incidents. It is likely that these people were going on a 'teerth' to Nanda Devi temple and had an accident/bad weather during the mountainous trek. It is this group which is called Roopkund_A that I will analyze in this post. So far, it is the only high coverage group of ancient DNA that we have from India.
What makes this group A so fascinating is that all these individuals are unrelated and are like a snapshot of the whole of India at that particular time (800CE). The sample size is also decent. The PCA below will explain.
PCA for Roopkund A
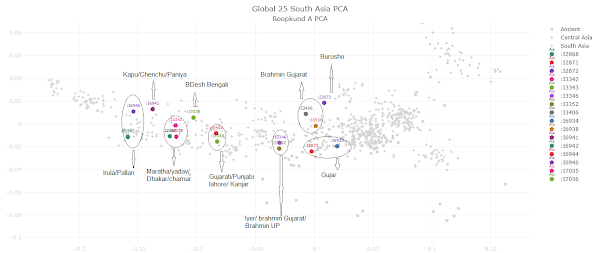 |
| 16 of the 23 A group samples, and note of which modern population is closest to each. Click to enlarge. |
All 23 Roopkund_A samples and best qpAdm models.
| Sample ID |
Gender |
Y HG |
p-Value |
I8726 |
Sintashta |
Irula |
WSHG |
Mongolia_N |
Tarim_EMBA |
| I3351 |
M |
J2b2a~ |
0.8420 |
31% |
34% |
34% |
|
|
|
| I6938 |
F |
- |
0.2296 |
27% |
29% |
38% |
|
|
6% |
| I2872 |
F |
- |
0.0772 |
41% |
28% |
31% |
|
|
|
| I3406 |
M |
J2a1a1b1 |
0.1210 |
47% |
26% |
15% |
4% |
10% |
|
| I6943 |
M |
J2b2a2b~ |
0.8163 |
37% |
24% |
39% |
|
|
|
| I2871 |
F |
- |
0.1623 |
44% |
23% |
33% |
|
|
|
| I3349 |
F |
- |
0.3871 |
31% |
22% |
47% |
|
|
|
| I3346 |
M |
E1b1b1b2a1a~ |
0.0375 |
32% |
22% |
47% |
|
|
|
| I6945 |
F |
- |
0.0743 |
10% |
20% |
70% |
|
|
|
| I6934 |
F |
- |
0.4091 |
54% |
18% |
28% |
|
|
|
| I6944 |
F |
- |
0.3330 |
21% |
18% |
61% |
|
|
|
| I3352 |
M |
R2a2b1b2b |
0.1488 |
48% |
15% |
37% |
|
|
|
| I7036 |
M |
R2 |
0.0670 |
18% |
14% |
59% |
|
10% |
|
| I3344 |
F |
- |
0.0945 |
45% |
11% |
44% |
|
|
|
| I3343 |
F |
- |
0.2011 |
28% |
11% |
61% |
|
|
|
| I3402 |
M |
H3b |
0.0067 |
41% |
11% |
33% |
|
15% |
|
| I6941 |
M |
R2 |
0.1235 |
8% |
10% |
74% |
|
8% |
|
| I3342 |
M |
H1a1a4b |
0.1230 |
29% |
9% |
53% |
|
10% |
|
| I7035 |
F |
- |
0.1260 |
20% |
6% |
74% |
|
|
|
| I2868 |
M |
H1a1b1 |
0.2126 |
20% |
6% |
74% |
|
|
|
| I3407 |
M |
H1a1a4b |
0.0880 |
15% |
4% |
81% |
|
|
|
| I6946 |
M |
R1a1a |
0.2905 |
|
|
100% |
|
|
|
| I6942 |
M |
R1a1a1b2a1a1a1f~ |
0.1593 |
|
|
100% |
|
|
|
I8726 = Indus periphery sample from Shahr-Sokhta Iran with least AASI (Onge like) component
Irula are Dravidian (also called Irula language) speaking tribals from Tamil Nadu
WSHG = Western Siberian Hunter-gatherers, samples found in Tyumen and Sosonivoy (Russia)
Mongolia_N = Mongolia North Neolithic samples with east Asian ancestry
Tarim_EMBA = Tarim basin samples from the Xiaohe culture 1900BCE
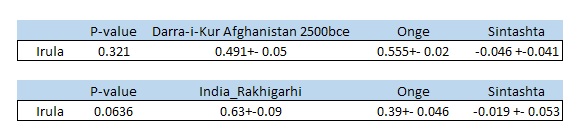
Irula itself can be modeled as (Rakhigarhi + Onge) or (Darra-i-kur_afghanistan + Onge) with -ve coefficients for Sintashta, or negligible noise level steppe ancestry considering std errors (results in folder link above). Additional proof for the absence of Steppe in Irula can be seen in Narasimhan et al supplement Table S84.
"Eight Modern Indian Cline groups (all tribal groups from southern India speaking Dravidian languages) in Table S 5 that are consistent with the proportion expected for an entirely ASI ancestry group in the sense that qpAdm analysis of these groups as part of the Indus Periphery Cline confirms that these groups fit without any Steppe pastoralist-related ancestry"
On performing statistical analysis on the above data, we can say that
- Irula component is statistically significantly associated with Y hg R1a at 99% confidence interval (independent t-test p-value = 0.0000107561) compared to non R1a. Click for the report.
- The Sintashta component is statistically significantly associated with the J2 y HG at a 99% confidence interval (independent t-test p-value = 0.00595690) compared with non J2. Click for the report. This is only possible if Sintashta autosomal ancestry was ultimately incorporated through steppe-rich females (as Sintashta male samples are all R1a). A similar result is seen in the Swat valley IA data (Narasimhan et al 2019).
- Y Hgs R2 and H are not statistically significantly associated with any component. Low sample size becomes relevant here as the null hypothesis could not be rejected like in the above 2 cases.
Above is my proof to all those who say and keep saying that there is a very high correlation between Steppe ancestry and R1a in modern Indians. The only proper aDna sample set we have from India shows exactly the opposite correlation.
On Y-Haplogroup R1a-L657
Y haplogroup can be determined by analyzing the Y chromosome of a male. The non-recombining portion of Y Chr is passed on only from father to son, and thus paternity can be determined. Once in many generations, one male undergoes one or many mutations in the Y Chr (eg C -> T, G->A, etc) and thus a subclade is born. This mutated subclade will now pass on through his son.
This is how R-L657 was born from Z93. (Brackets denote ISOGG notation)
R>R1>R1a>R1a1>R1a1a>R1a1a1>R1a1a1b>
> Z93(R1a1a1b2)> Z94(R1a1a1b2a)> Y3(R1a1a1b2a1)> L657(R1a1a1b2a1a)
Sample id I6942 is the only R-L657+ (+ means DERIVED or presence for the particular mutation, - means ANCESTRAL or absence of that mutation) found in ancient DNA so far. R-L657 is that subclade of R1a which is now common in the Indian subcontinent and the middle east, but not found in modern Europe or ancient Europe/steppe. The ISOGG name for it is R1a1a1b2a1a. Technically, I6942 is + for R-Y5 (R1a1a1b2a1a1a1~) and R-Y928 (R1a1a1b2a1a1a1f~) which implies that it is + for L657 as well.
It is safe to say that R-Y3 & R-L657 and their subclades were born in the Indian subcontinent given their absence in modern and ancient steppe and Europe (except Indian immigrants and the Romanis). I have always maintained that given that the formation and spread of R-Y3 is 2600 BCE and R-L657 is 2200 BCE respectively (YFull), that their paternal ancestors (Z93 and Z94) were somehow present in the Indian subcontinent but without the presence of autosomal steppe ancestry which enters Indian subcontinent only post 1500 BCE.
How did R-L657 reach India?
Here, it is worth noting that the oldest Z93 samples have been found in Fatyanovo culture, Russia dating to 2500 BCE. So it is likely that this Z93 did enter India around the same time from Russia, however, it would not be accompanied by any noticeable change in autosomal ancestry. Autosomal ancestry is that which recombines and is present in Chr 1-22 and is passed 50-50% from both parents to the child. Steppe autosomal ancestry only enters India post-1500 BCE.
The other option of course is that India was the source of R1a to Europe, to prove which we need tonnes of ancient DNA which we don't have. Europe also has ancient samples of ancestors of Z93, like Z645 and M417 and so on, which makes the European origin of R1a much more likely. So I will not consider this option yet, although it's plausible. There can always be haplogroups that reach multiple places due to some travel-savvy ancients :)
This has happened before, we see y hg J2 and J1 (and subclades) from Iran, the Caucasus, and SC Asia neolithic in European samples in Karelia_EHG and Austria LBK, Hungary_Sopot without any apparent autosomal ancestry from the source regions. Autosomal ancestry from a parent can dilute to ~0% easily in 7 generations if the sons end up marrying local women (50%>25%>12.5%>6.25%>3.125%>1.5%>~0) ie 150 years.
Some make the argument that the lack of R1a in Shahr Sokhta, BMAC, Turan Eneolithic proves that R1a was absent in this region. To which my response is that these regions saw population turnover starting 4th mill BCE or even earlier by a population like Tepe Hissar/Seh Gabi to the tune of >60% and a resulting Y HG turnover. This is the reason why west Asian Y haplogroups like E1b and G2 appear in the record in the regions adjoining NW India. This turnover never happened in the Indian subcontinent as Indians lack this Anatolian heavy component. So I don't consider this evidence very strong, more data is needed.
SUMMARY
1. Sintashta component not correlated with R1a in RoopkundA samples, whereas Irula component is.
2. J2a/J2b significantly correlated with higher steppe ancestry in RoopkundA samples, steppe ancestry mediated through females.
3. I discuss possibilities regarding R-L657 reaching India.
With this, I present the results of the L657 poll, I guess it's clear that the Indian subcontinent won this one.
Also, read



if i remember correctly, 50% of iranian zoroastrians and 50% priests were haplogroup J. Which is interesting considering above analysis.
ReplyDeleteterrific analysis.
ReplyDeleteconvincing response to Narasimhan's recent question in a CH room on why R1a-L657 has not appeared in central Asia aDNA, if its origin is from India/SC Asia.
ReplyDeleteCan u explain in detail??
Delete> The other option of course is that India was the source of R1a to Europe, to prove which we need tonnes of ancient DNA which we don't have. Europe also has ancient samples of ancestors of Z93, like Z645 and M417 and so on, which makes european origin of R1a much more likely.
ReplyDeleteI am excited for Niraj Rai's paper on basal R1a lineages in India. It will bring new light onto this topic.
Yea, i have been waiting a long time for that.
DeleteExcellent Post!
ReplyDeleteTwo things stood out for me. You have written that the Roopkund_A group is an excellent stand-in for the India of 700-900 CE. And 23 individuals is a very good sample size.
This is effectively a double blind study. The participants don't know anything about the analysis they are being subjected to (long dead!!) and the researchers don't know anything about the participants. In most modern studies, there is a questionnaire accompanying DNA studies which allows for the researcher to indulge in bias.
Is it possible that you can select a random sample of 23 modern Indians (randomisation is key) and do a drift analysis of haplogroups and autosomal components??
What would that accomplish?
DeleteIt can be done using eurogenes g25 coordinates and vahaduo. Just don't use the simulated AASI coordinates famously used in anthrogenica as source. They're faulty.
Ok, thanks! I wanted to know if it is possible to establish the degree of admixture into India in the last 1200 years.
ReplyDeleteNo additional admixture into modern indians for the large part as compared to roopkund samples.
DeleteExcellent analysis 👏.
ReplyDelete"Above is my proof to all those who say and keep saying that there is a very high correlation between Steppe ancestry and R1a in modern Indians. "
ReplyDeleteI believe the people who make this correlation, do so in this sense of
ancient migrations not present day frequencies. We easily know how easily autosomal ancestry can change by few generations of intermarriage without affecting Y-chromosome.
A person outcasted from a caste, who is later taken by some other low caste/tribal group, lets call them X, his descendants can easily become X like in a few generations autosomally.
R1b-V88 is found in many african ethnicities/tribes, *some* of which barely have any "west eurasian" admixture. Based on this would we claim that R1b-V88 arose in central africa ?
My reasoning for L657 birth in India is that it's absent in ancient records of steppe and in modern Europeans. Not because it was found in roopkund.
ReplyDelete@Vasistha
ReplyDeleteIt is safe to say that R-Y3 & R-L657 and its subclades were born in the Indian subcontinent given its absence in modern and ancient steppe and Europe (except Indian immigrants and the Romanis). I have always maintained that given that the formation and spread of R-Y3 is 2600 bce and R-L657 is 2200 bce respectively (YFull), that their paternal ancestors (Z93 and Z94) were somehow present in Indian subcontinent but without the presence of autosomal steppe ancestry which enters Indian subcontinent only post 1500 bce.
Modern and ancient distribution of Y3, Y2 and even L657 suggests to me that they or at least Y3 and Y2 appeared somewhere in South Central Asia rather than South Asia (the Indian subcontinent).
https://www.yfull.com/tree/R-Y3/
@onur
ReplyDeleteIndia is severely undersampled in yfull. hopefully the 10k modern sample research will be published soon. the preview shows lots of Y3 and Y2 in the dataset.
The Alai Nura 300CE R-Y3 is during the time buddhists were travelling through that route to and from china.
ReplyDelete@Vasistha
ReplyDeleteIt would be good if at least some of those new academic male samples are uploaded to YFull.
@Vasistha
ReplyDeleteSo you think Z93 and Z94 formed somewhere in Eastern Europe, L657 and its immediate ancestors Y3 and Y2 formed in the Indian subcontinent and Z2124 again formed somewhere in Eastern Europe. What about Y40? Where do you think it formed?
Yea Z2124 definitely not Indian, but Y40 looks like it is.
ReplyDeleteNeed more clarity on these commonalities of R1a between middle East and South Asia, are they only recent migrants or is there bronze age connection? Could also be turkic speakers carrying SC asian Y hgs post Buddhism to middle East.
@Vasistha
ReplyDeleteThe notion that R/R1a haplogroups intruded into India only with the arrival of the Steppes autosomal component is a very good candidate for a null hypothesis.
And I think just modern Indians DNA data can provide sufficient parametric conclusions to accept it or reject it. No need for aDNA.
It is the alternative hypothesis that is difficult to frame. I have 3 candidates -
1. Steppes autosomal contribution arrived via females.
2. Steppes autosomal contribution arrived via males but not bearing R/R1a haplogroups.
3. Steppes autosomal contribution indeed arrived via males bearing R/R1a haplogroups but they were already present in India.
The requirement of aDNA only arises to pick a correct alternative hypothesis. Any thoughts on this?
When we say steppe males, we only mean R1a males as steppe_mlba is 100% R1a. So #2 is not needed.
ReplyDelete@Vasistha
ReplyDeleteAhh....thanks. Then your L657 conjecture is more than a sufficient proxy for Option 3. The absence of it in the aDNA record of Steppes while its occurence in modern Indians is remarkable.
Option 1 is in as there is sufficient textual evidence (Mahabharata) of women from the Northwest marrying into Indian royal lineages (Gandhari, Madri etc). Most secular evidence points to the MbH taking place in the 1400 to 900 BCE period. It curiously dovetails with the genetic evidence for the autosomal intro.
Waiting for Niraj Rai's aDNA from BRW/OCP/PGW cultures.
"Option 1 is in as there is sufficient textual evidence (Mahabharata) of women from the Northwest marrying into Indian royal lineages (Gandhari, Madri etc)."
DeleteBut how would you explain decent persence of steppe autosomal ancestry among non-royal groups like jats, rors, ahirs. Even in the gangetic plains, non-dwija middle castes of gangetic plains had around ~15%-16% steppe ancestry autosomally in narasimhan's paper but based on few mtDNA reports i have seen, i barely saw any steppe mtDNA lineage among these castes .
"Waiting for Niraj Rai's aDNA from BRW/OCP/PGW cultures."
He claimed last year that his team's findings would be out this year but the year is nearly gone .
Major Y-Haplogroup of Jats
DeleteHaplogroup L (36.8%)
Haplogroup R (28.5%)
Haplogroup Q (15.6%)
Haplogroup J (9.6%)
3 of these are deeply rooted in India with Q arriving during Hunnic Turkic and Mongol invasions.
I am no expert but western Scythians descended from Andronovo people and carried Y-Haplogroups like R1a1a1b2a2a. Many Scythians settled in India and formed pre Islamic lordships in northern and western India.
It's highly unlikely they all got slaughtered in Turkic invasions.
Darius in one of his inscriptions dated ~490bce mentions Hindush next to Saka so it's highly likely Scythian were present in India much before they carved out an empire in India.
Q1b is present in loebanr 1300 bc and Q1* BASAL,Q1a1,q1a3,q1b is present among zoroastrians,gilaks,etc.
DeleteIt's not from Huns or Turks.
It's from hotu ANE,Same population that gave q1b to ashkenazi and other jews
@Vasistha
ReplyDeleteYea Z2124 definitely not Indian, but Y40 looks like it is.
Need more clarity on these commonalities of R1a between middle East and South Asia, are they only recent migrants or is there bronze age connection? Could also be turkic speakers carrying SC asian Y hgs post Buddhism to middle East.
Here is the YFull page for Y40:
https://www.yfull.com/tree/R-Y40/
And here are the public people grouped under Y40 on the FTDNA R1a Project (these are the public ones, so I can share their results with no hesitation):
https://drive.google.com/file/d/1VPOzPbCbrMYHvqGDc3nQNR6ZgcGWm5of/view?usp=sharing
Based on these, where Y40 itself formed seems open to speculation, the three of its four main branches might well have formed in the Indian subcontinent or somewhere in Southern Central Asia, but the YP294 branch seems unrelated to South Asia, not being found there and only being found in West Asia and parts of Europe (as far as Germany and Italy), at least based on YFull and FTDNA (have checked the country reports of the Y-DNA haplotree of FTDNA too). YP294 formed about 4500 years ago according to YFull like the other three main branches and Y40 itself.
Y40 and especially its YP294 branch are found in many West Asian peoples (whether Muslim or Christian) and even some European peoples with no genetic Turkic connection. Also I have never seen Y40 in any Central Asian Turkic people or in populations such as Tatars and Bashkirs. Add to that the old ages of Y40 branches in West Asia and Europe, we can safely infer that Y40 has nothing to do with the Turkic migrations, it seems connected to the Indo-Iranic migrations and maybe also some other IE migrations.
Thanks onur
Delete@onur
ReplyDeleteWe do see in archaeo and animal genetic record a steep increase in zebu and asian elephant in Syria starting 2000bce.. likely connected to this R1a variant.
@Vasistha
ReplyDeleteWe do see in archaeo and animal genetic record a steep increase in zebu and asian elephant in Syria starting 2000bce.. likely connected to this R1a variant.
The timing makes sense. The said animal migrations might be connected to the migrations of L657 and its Y3 and Y2 predecessors to West Asia. But I am not sure if Y40 came to West Asia with the same migrations as Y40 has not been found in ancient DNA so far to my knowledge and it is rarely found in modern DNA and thus hard to trace to the past eras.
Indeed, y40 needs to be found in adna
ReplyDeleteWill address Laziridis's old tweet from jan 21 here with relation to this topic
ReplyDeleteHe says:
"- L657 is downstream from Z93.
- Roopkund samples are thousands of years after the migrations of Z93 bearers.
Why do you focus on "no steppe component" in medieval samples from India and not on "no South Asian component" in 3rd/2nd millennium BC steppe samples?"
Quite simplistic takes as expected.
1. L657 is indeed downstream of Z93 (which is most likely from russia).. however,
L657 formation is pre 2000bce. Given that it is not found in europe then and now, it means L657 was born to one man inside subcontinent (as two people cannot independently have same mutation), rather than . Can the progenies of this one man change the language of a europe sized country without leaving trace of existing language?
Given that the steppe forefather of this L657 came pre 2000bce without much lasting autosomal ancestry, it is very likely that the correlation between autosomal and y Hg got broken by dilution. ie. the autosomal steppe ancestry got diluted whereas the non recombinant Y chr passed down as is.
There was the major steppe autosomal wave which happened later post 1500bce which we see in Swat, this one mediated through steppe women.
2. there need not be south asian component (AASI) that we need to look for. That is a strawman. Darra-i-kur 2500bce afghanistan does not have any aasi or anatolian. it is simply IndiaN + WSHG.
I have shown in my steppe eneolithic post the high possibility that the contributing ancestry was from east of caspian..
793 new samples from england and europe in new paper.
ReplyDeletehttps://www.nature.com/articles/s41586-021-04287-4
all samples added to https://a-genetics.blogspot.com/2021/12/adnamap.html
Mitanni vs Indo Aryan
ReplyDelete( Punjabi/Sindhi/Koshur/Sanskrit)
aika - Ikk - akh - hiku - eka
tera - Tin - tre - ṭī - tri
panza - Panj - Pancs - pañja - pañcan
satta - Satt - sath - sata - sapta
na - No - nav - nava - nava
Mitanni vs Iranian
(Kurdish/Persian/Pashto/Blauchi)
aika - yek - yek - yəw - yak
tera - sê - se - dre - sy
panza - pênc - panj - pinzjə - panch
satta - heft - haft - ûwa - hapt/heft
na - no - noh - nəh - nuo
Camel motif from Sanauli was single humped compared to double humped camel of BMAC. Along with peacock motif it indicates that Sanauli people were locals and not from BMAC or Andronovo as some idiots claim.
ReplyDeleteCHG and IranN were related groups though I haven't heard anyone say that. Instead they say CHG was different from IranN due to isolation for thousands of years but I don't know what they mean because they at the end of the day were formed from same group of ANE and .....I don't remember the name right now something starting with D.
ReplyDeleteIf CHG was PIE signal then in my opinion there is every possibility that IranN also spoke PIE or dilect related to it.
There are Indo-Aryan sounding words in Mesopotamia like Aratta Anu etc attested far before Mitannis Kassites and Hittites appeared in Near East.
Aratta being one of special interest as it finds mention in our texts as well. Aratta as mentioned in Sumerian legends was somewhere between eastern Iran and Indus.
It is a fabulously wealthy place full of gold, silver, lapis lazuli and other precious materials, as well as the artisans to craft them.
ReplyDeleteIt is remote and difficult to reach.
Afghanistan Shortugai was source of lapis Lazuli. Shortugai was Harappan site.
The lord of Aratta, named En-suhgir-ana (or Ensuhkeshdanna)
Suhgir or Suhkesh are both Indian sounding names ^_^
R* was found in malta-boy in Eastern Siberia 25000BP
ReplyDeleteIf IranN had half descent from ANE male and IranN was major ancestry component of people of Indus valley civilization wouldn't Y-Haplogroup R be present in North West India from Neolithic time?
If I am not wrong R2a has been found from IranN rich Ganj Dareh sample and one Indus valley periphery sample was also from R Y-haplogroup.
Was Y-Haplogroup L and H AASI specific?
In case this ANE source was female mediated then it was possibly the source of Haplogroup U?
The other half of this IranN ancestry was possibly a source of Y-haplogroup like J if it was mediated through male.
Or Probably I don't know anything and making stupid assumptions and claim which will be clarified only if somebody like you answers it.
On YouTube I came across 2 Indians; one with Y-Haplogroup C-M130 from Gujarat and one with Y-Haplogroup R-Y7 from Punjab.
ReplyDeleteWould like to know about their possible origins from you. Are they in any way linked to Hunnic and Scythian migrations/invasions into India?
Steppe ancestry was introduced into India both ways.
ReplyDeleteFirst through females post 2500bce and then through males post 1000bce and this kept coming as late as 500ce;
Parathians
Scythians
Kushanas
Hepthalites
There was also large scale trade going on through silk route. Can't underestimate such an impact.
Don't underestimate the power of Polygamy. 1 male is more than enough to reproduce 60 kids in his life time. As per our legends Daksha had 60 daughters.
ReplyDelete1 R1a male is enough to reproduce 30 males carrying R1a and 50-50 Steppe to non-Steppe ancestry and 30 females with same ancestry in a very short time.
Haven't this been found to be true in the recent paper which details about 1 male with 4 wives and 27+ member family from 3500bce England.
There is a poor couple where I live who have reproduced 11 kids. 11 from just 1 wife.
You don't need invasions and migrations to explain it. But their real objective is to prove that Harappans weren't indo-Aryans hence this 200 year non sense.
As per my hypothesis:
ReplyDeleteAndronovo can't be proto-indo-Iranian. There is 0 literary evidence to make such an absurd claim. At best they could be proto-Scythians.
Post 1000bce we see the appearance of Persian speaking groups in South East and Northern Iran.
Western Iran was still multilingual with Elamite and Assyrian speakers dominant in the region.
Indo-Aryan Mitannis survived as late as 1200bce after which they either disappeared or got assimilated or possibly went North of Caucasus taking their Aryan language into balto-slavic region.
This group of people can't have transformed into Iranic speaking tribe.
Balto-Slavic languages are attested in common era whereas as Mitanni Aryan and Mycenaean Greek have bronze age attestation.
ReplyDeleteThe difference in dates but similarities in language family only point to the South-North direction.
Much of Iron Age before common era people of eastern Europe were Aryan speakers of Iranian type. More than enough to explain the similarities between balto-slavic and Aryan languages.
Scythian dominated Eurasian steppes for 2000 years of you consider Andronovo as proto-Scythian which I am sure can be proved through genetics.
I read somewhere that Poland could be the homeland of Slavic languages. But I can't vouch for the authenticity of this claim.
Chg are iranN don't form a clade.. ie they don't have a common ancestor. They're formed from different proportions of the same components.
ReplyDeleteWrt mitanni, yes the language sounds similiar to sanskrit than iranic. Likely brought to Syria along with the evidence we see for zebu cows and indian elephants post 2000bce.
Yes R2 is iranic/indian in origin.. we don't see it elsewhere. R1 would have formed closer to R2. Or in the area between ANE and Iran/india.
Hap C has widespread occurence, but east asia/Siberia is likely origin.
R-y7 is downstream of L657, so its the local variant of R1a.
"First through females post 2500bce and then through males post 1000bce and this kept coming as late as 500ce;
ReplyDeleteParathians
Scythians
Kushanas
Hepthalites"
All of the scythians had extra east asian ancestry which we can't detect pan India yet.. need more 500bce - 200ce samples to see the impact of this scythian ancestry
Where did MongolianN ancestry in 5 Roopkund samples come from?
DeleteSome one once shared their ancestry on Twitter
ReplyDelete56.0 IRN_Shahr_I_Sokhta_BA3
29.4 Yamnaya_RUS_Samara
5.6 IRN_Ganj_Dareh_N
4.2 Anatolia_Barcin_N
3.2 Levant_PPNB
1.0 Han
0.4 Jarawa
0.2 WHG
Your take.
That 1.0% Han got me interested.
ReplyDelete"Around 2700 BCE, at the end of Phase 7, most of the Shahr-i-Sokhta city was destroyed by a fire, particularly the Eastern Residential Area and the Central Quarters showed "rooms with burnt plaster, filled with ash and burnt remains of roof beams."
So far, this seems to be the only proof of any invasion in the region of Iran and NW India, although it could also be natural accident. This is also the time when SiS sees harappan contacts (and the indus periphery samples proving the connection). In that sense, the connection of Agni taming the godless is interesting.
"56.0 IRN_Shahr_I_Sokhta_BA3
29.4 Yamnaya_RUS_Samara
5.6 IRN_Ganj_Dareh_N
4.2 Anatolia_Barcin_N
3.2 Levant_PPNB
1.0 Han
0.4 Jarawa
0.2 WHG"
This is a bad model, ignore it. Its a mix of neolithic & BA sources. Should have used all BA sources with han and jarawa if required. SiSBA3 is low coverage and should not be used unless no choice. ganj_dareh is 9000yrs old, same for levant and barcin. whats the point?
The mongolian ancestry is a stand in for any sort of added east asian ancestry - you see it in bangladeshis, and UPites (extra pahadi ancestry).
Im open to the idea of scythian ancestry in India, need more data for that.
Thanks ^_^
Delete"Was Y-Haplogroup L and H AASI specific"
ReplyDeleteH can be AASI specific but L can't imo. This faulty connection of L with AASI comes from L1a1(L-M27) presence among south indian castes(mainly along the western part of south) but L's another clade L1a2(L-M357) is a north west south asian lineage with it being predominant among Jats(both hindu and sikh jats), Rors etc. This lineage has also been found in the BMAC region and L1a has also been found in samples from chalcolithic armenia(with no AASI).All these make evidences make it highly unlikely for L to be AASI specific. The L1a1(L-M27) in south indians today, imo, comes from the farming pops who moved to south from north.
"Major Y-Haplogroup of Jats
ReplyDeleteHaplogroup L (36.8%)
Haplogroup R (28.5%)
Haplogroup Q (15.6%)
Haplogroup J (9.6%)
3 of these are deeply rooted in India with Q arriving during Hunnic Turkic and Mongol invasions."
@AshishKaull, this seems to be taken from David Mahal's paper, now his paper doesn't expand on the subclades of L, R and J found among Jats which makes his paper useless in my eyes. Based on seeing Y-HG data of many jats of different clans aggregated from various testing labs, here are the following observations:
- Nearly all of Jats L belong to L1a2 or L-M357 which is different from the predominant L clade found among south indians( L1a1 or L-M27). This L-M357 is more of a NW south asian clade with presence in afghanistan and central asia. Some of Swat valley iron age samples had this clade.
- Nearly all of Jats R belong to R1a-L657 with a few belonging to R2 and a very small minority having R1b
Of course, it depends upon what you mean by 'deeply rooted in India', if you mean neolithic and before, then I am not sure if either L-M357 or R1a-L657 can be thought to be present in south asia since as early as that.
There is a 3 week old video of Niraj Rai where is said;
Delete01 2 to 2.5 times diverse than Europe (R1a1) likely to be present within South Asia 15000BP-10000BP
02 Tribal population groups in South India have this up to 30%
03 At least 50 mutations
04 Harappa era samples from Kalibangan, Lothal, Dholavara
05 Interesting sample from Burzhom Neolithic Kashmir
06 No central Asian or Eastern european component till 1300bce ( I wonder if he means no such ancestry from ancient samples taken from modern Indian territory) because we do find Swat valley samples post 1200bce with small portion of steppe ancestry. If there really was a large scale intrusion we would have found high to complete steppe male mediated ancestry which to this day evades us.
Delete07 large scale central Asian add mixture post 3rd century bce
Fall of BMAC (1700bce) could have caused backward migrations of descendants of steppe Harappans born of steppe mothers.
15% Q is interesting
ReplyDeletehttps://www.tehrantimes.com/news/468385/Elamite-clay-tablet-unearthed-in-mysterious-Burnt-City
ReplyDelete"Called “Shahr-e Sukhteh” in Persian, the UNESCO-registered Burnt City is associated with four rounds of civilization, all mysteriously burnt down by catastrophic sets of fire."
all 4 stages burnt down by fire, sounds like war all right.. Then this is probably the Aryas unleashing Agni on the Dasyus lmao.
Interesting!
DeleteShahr-e Sukhteh ain't that far from helmand river which some scholars identify with Haraxvati/Saraswati.
But as far as I know Helmand ends in a lake where as Rig Veda 7.95.2 says from Mountain to Ocean.
DeleteSo equating the two is probably wrong. More so Avestan name for Helmand is Haētumant.
I wonder what is meant by this
"For thee, O Blest Sarasvatī, Vasiṣṭha hath here unbarred the doors d sacred Order."
Hi Vasistha,
ReplyDeleteI have a question. Did the Sredny Stog sample found in Alexandria, Ukraine belong to Y-HG R1a-Y3 ? Was it positive for Y3 ?
https://www.biorxiv.org/content/10.1101/2021.11.04.466891v1.full
ReplyDeletePerle Guarino-Vignon et al. Is an interesting paper but I just get lost mid way through it.....
I have read it multiple times but ....x_x
Rig Veda 7.6.5
ReplyDeleteHim who brake down the walls with deadly weapons, and gave the Mornings to anoble Husband,
Young Agni, who with conquering strength subduing the tribes of Nahus made them bring their tribute.
@AshishKaull,
ReplyDelete"01 2 to 2.5 times diverse than Europe (R1a1) likely to be present within South Asia 15000BP-10000BP"
What does diversity here mean ? Anyways, concluding the time since any haplogroup has been present in an area without ancient DNA doesn't seem reliable.
"02 Tribal population groups in South India have this up to 30%"
He is probably talking about Chenchus, a small tribal pop who seem to have founder effect of R1a though not sure if their R1a is of L657 type
"06 No central Asian or Eastern european component till 1300bce ( I wonder if he means no such ancestry from ancient samples taken from modern Indian territory) because we do find Swat valley samples post 1200bce with small portion of steppe ancestry. If there really was a large scale intrusion we would have found high to complete steppe male mediated ancestry which to this day evades us."
He is probably alluding to the ancestry of PGW(painted grey samples) and immediate successor cultures of PGW in the upper gangetic plains. Wonder how PGW people turn out to be autosomely ?
"07 large scale central Asian add mixture post 3rd century bce"
This is new information ! Can you give the link of the video ?
"Fall of BMAC (1700bce) could have caused backward migrations of descendants of steppe Harappans born of steppe mothers."
How does this explain the very high bronze age steppe ancestry of groups like Rors ? mtDNA haplogroups of Rors can been seen at page no 28 of https://ars.els-cdn.com/content/image/1-s2.0-S0002929718303987-mmc1.pdf
The paper claims that 37.5% mtDNA of Rors are 'west eurasian' however, these 'west eurasian' mtDNA lines also consist of U7(7.7%) and W(7.7%) which are more like central asian/iranic and NW south asian origin. Subtracting the %s of U7 and W, we get AT MOST ~23% mtDNA haplogroups which CAN be bronze age steppe lineages while on the autosomes, Rors show around 35% bronze age steppe related ancestry. What's the case here ?
Weren't Indus valley people West Eurasian shifted since Neolithic times? Rakhigarhi female belonged to U2b2. Don't remember the Haplogroups of IV_perephary samples.
DeleteMtdna U appears to be common among Andronovo people.
I think there was this old paper as per which R1a M17 was found at high frequency among South Indian tribal groups. R1a M17 is also found in high frequency among Rors.
DeleteAs far as I know Chenchu have no steppe ancestry and phenotypically they are quite distinct from Rors.
That's quite interesting ^_^
"I think there was this old paper as per which R1a M17 was found at high frequency among South Indian tribal groups. R1a M17 is also found in high frequency among Rors."
DeleteThat was a paper by a Kashmiri researcher, and you are probably talking about Saharia(a tribal group in central india) and Chenchus.
"Chenchu have no steppe ancestry and phenotypically they are quite distinct from Rors.
That's quite interesting ^_^"
Tbh, i don't find anything interesting here, as vasistha wrote,- autosomal ancestry can completely dilute within 7 generations of intermarriage so even historical era introgression of R1a among chenchus can be completely wiped out of its autosomal signatures.
"Rakhigarhi female belonged to U2b2.Mtdna U appears to be common among Andronovo people."
There are two variants of U2 - one is india/south asia specfic variant which are collectively called U2i(U2b and U2c are in umbrella) and another is Europe specific variant called U2e, the andronovo samples were U2e.
@tim
ReplyDelete"I have a question. Did the Sredny Stog sample found in Alexandria, Ukraine belong to Y-HG R1a-Y3 ? Was it positive for Y3 ?"
1. I6561 has been redated to 2000bce from 4000bce or so earlier.
2. I6561 is Z93+ for sure, with no reads at Z94/Z95 but derived C-->T at Y3 but ancestral at Y2.
Since this sample is not corrected for aDna damage at ends of the dna fragment, this C--->T is highly likely to be a deamination error, the most common form of error in damaged aDna.
in last version of harvard anno file, it was labeled QUESTIONABLE (damage=0.028), in this one it is labeled pass, damage=0.028 for some reason.
in short a lone C-->T call in a damaged sample is not good enough. the sample is simply z93+
Link to rai video plz.
ReplyDeletehttps://m.youtube.com/watch?v=_4L-l8nJA24
Delete"No central Asian or Eastern european component till 1300bce"
ReplyDeleteNiraj rai has seen results of 1300bce samples, probably north indian, thats why he is saying this.
https://m.youtube.com/watch?v=_4L-l8nJA24
ReplyDeleteRor population is very small I wonder how they maintained their ancestry. We Kashmiri Pandits number same but endogamy doesn't appear as strict as Rors despite living in a much more isolated location.
700k Rors 5000 odd Kalash and 100 odd Sentinelese do appear out of place in a 1800 million strong South Asian population.
I was given a spread sheet by someone on another forum;
Jats do appear to have high CHG-NE Euro like ancestry but I have no idea where Rors stand as per Razib khan's/Harappa world database.
Seems like Not all groups and castes originated from one homogenous event. Taxali and other Indus valley territories fell into the hands of Iranians by 535bce.
Indians formed a unit in Persian army and were part of various invasions in Greek and sourounding areas.
"Ror population is very small I wonder how they maintained their ancestry."
DeleteThere is an another group in NW india which has a very large population but very similar to Rors in ancestry - Jats. In fact, most of the clans names of jats and rors are exactly the same so one wonders when this split of identity between Jats and Rors occurred.
While Y-HG pattern of both Rors and Jats are similar (two primary Y lineages dominate both - L-M357 and R1a-L657), i don't have good data on jats mtDNA lineages. If Jats indeed turn out to have good frequency of steppe mtDNA lines then the theory of bronze age steppe ancestry coming primarily from females will get a boost.
@ashishkaul
ReplyDeleteneed more samples from india in 1 study which analyzes everyhting together. chenchu by itself doesnt mean anything.
Yes
DeleteI always favoured R1a L657 to be in India in the 3rd milennium BCE. This has a lot of implications for the date of Rig VEda and Indian history in general.
ReplyDelete